Developmental Biology Program
The Mary Baylies Lab
Research

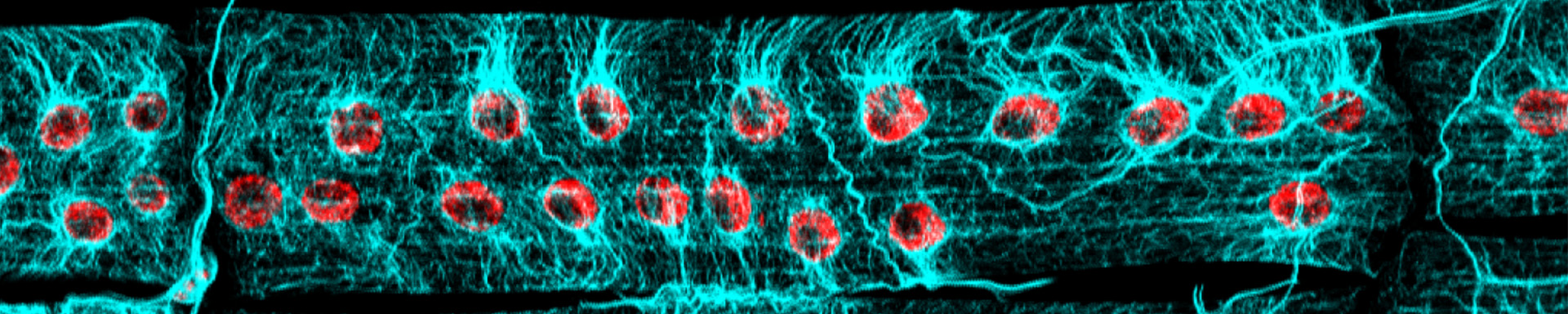
Developmental biologist Mary Baylies studies the mechanisms that form and maintain muscle both during normal development and in disease.
Research Projects

Featured News



Publications Highlights
Windner SE, Manhart A, Brown A, Mogilner A, Baylies MK. (2019.) Nuclear Scaling Is Coordinated among Individual Nuclei in Multinucleated Muscle Fibers. Dev Cell. Apr 8;49(1):48-62.e3. Featured as a cover image and in a Preview.
People

Mary Baylies, PhD
- The Baylies laboratory studies the mechanisms that form and maintain muscle both during normal development and in disease.
- PhD, The Rockefeller University
- [email protected]
- Email Address
- 212-639-5888
- Office Phone
- Download CV
- PDF File
Members
















































Achievements
- Identified that global, regional, and local inputs contribute to nuclear size regulation in multinucleated skeletal muscle fibers.
- Demonstrated that properly placed nuclei are critical for healthy muscle function.
- Discovered that microtubules are critical for the movement and positioning of nuclei in the multinucleated skeletal muscle cell.
- Developed Drosophila models of muscle wasting in cancer cachexia and aging.
- Identified how actin filament dysregulation contributes to skeletal muscle dysfunction in models of nemaline myopathy.
- Developed a Drosophila model for metastasis in alveolar rhabdomyosarcoma, a type of soft tissue cancer most often seen in children.
- Described how nuclear polarization occurs during myonuclear movement in vivo.
- Completed the first spatial and temporal characterization of an F-actin structure that forms at the myoblast fusion site.
- Identified transcription factors and chromatin regulators that are critical for muscle identity.
- Identified 3-D arrangement of myoblasts that occur during muscle specification and morphogenesis.
- Discovered signal transduction pathways (Notch, Wnt, BMP, and RTK) and transcription factors that are critical for muscle identity.
- President of the Society for Muscle Development (2009-2013).
- Frederick Adler Chair for Junior Faculty (1997-2003).
Open Positions
To learn more about available postdoctoral opportunities, please visit our Career Center
To learn more about compensation and benefits for postdoctoral researchers at MSK, please visit Resources for Postdocs
Get in Touch
-
Lab Head Email
-
Office Phone
-
Lab Phone
-
Lab Fax
To learn more about Postdoc compensation and benefits at MSK, please visit https://www.mskcc.org/education-training/postdoctoral/resources-postdocs/compensation-benefits-resources
Disclosures
Members of the MSK Community often work with pharmaceutical, device, biotechnology, and life sciences companies, and other organizations outside of MSK, to find safe and effective cancer treatments, to improve patient care, and to educate the health care community. These activities outside of MSK further our mission, provide productive collaborations, and promote the practical application of scientific discoveries.
MSK requires doctors, faculty members, and leaders to report (“disclose”) the relationships and financial interests they have with external entities. As a commitment to transparency with our community, we make that information available to the public. Not all disclosed interests and relationships present conflicts of interest. MSK reviews all disclosed interests and relationships to assess whether a conflict of interest exists and whether formal COI management is needed.
Mary Baylies discloses the following relationships and financial interests:
-
French National Research Agency
Professional Services and Activities (Uncompensated)
-
Howard Hughes Medical Institute
Professional Services and Activities
The information published here is a complement to other publicly reported data and is for a specific annual disclosure period. There may be differences between information on this and other public sites as a result of different reporting periods and/or the various ways relationships and financial interests are categorized by organizations that publish such data.
This page and data include information for a specific MSK annual disclosure period (January 1, 2024 through disclosure submission in spring 2025). This data reflects interests that may or may not still exist. This data is updated annually.
Learn more about MSK’s COI policies here. For questions regarding MSK’s COI-related policies and procedures, email MSK’s Compliance Office at [email protected].


