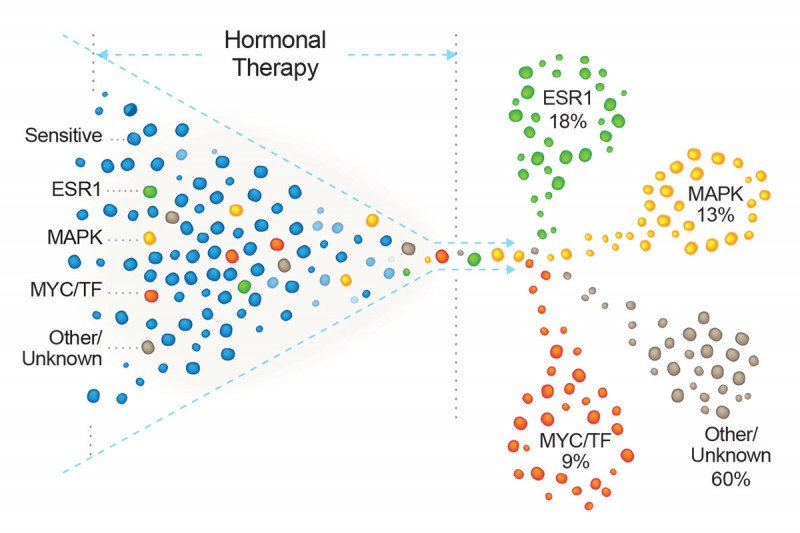
A study of hormone-resistant breast cancer tumors finds that they sort into one of four different categories based on their genetic mutations. Image credit: Cancer Cell
Estrogen receptor (ER)–positive breast cancer is the most common type of the disease, accounting for 70% of all cases. Hormonal therapies designed to starve the cancer of estrogen or block its receptor have greatly improved outcomes for women with these tumors, preventing recurrence and delaying disease progression.
Unfortunately, for those with metastatic disease, resistance to hormonal therapies eventually emerges in essentially all cases, as cancer cells evolve other ways to support their continued growth. Ultimately, resistance is what leads to progression and death.
Now, Memorial Sloan Kettering researchers have combed through more than 1,500 ER-positive breast cancer tumors to establish a taxonomy, or catalog, of resistance-associated genetic changes. The results, published today in the journal Cancer Cell, identify previously unknown drivers of resistance and portend changes in the way that ER-positive breast cancer is treated.
Familiar and Unfamiliar Genetic Suspects Driving Resistance
In the study, a team of researchers led by MSK’s Physician-in-Chief José Baselga looked at DNA from 1,501 ER-positive tumors from 1,364 women being treated at MSK. About half of the samples came from women who had not yet received hormonal therapy, and the other half from women who had already received this treatment. The majority of the tumor samples were from metastatic sites of disease, making this the largest clinical–genetic study of hormone resistance in patients with metastatic ER–positive breast cancer to date.
Genetic sequencing of the tumors was performed using MSK-IMPACT™, a test that looks for mutations in 468 cancer-associated genes.
From previous work done at MSK, it was known that mutations in ESR1, the gene that codes for the estrogen receptor, are a main driver of resistance to hormonal therapy in people with breast cancer. Normally, the estrogen receptor requires estrogen to bind to it before it can turn on genes that promote tumor cell growth. But the mutated versions no longer require this binding. Cells with these mutations are therefore able to evade therapies designed to starve the cancer cells of estrogen. The most common of these drugs, called aromatase inhibitors, block the enzyme that makes estrogen.
In the analysis, the researchers found ESR1 mutations in 18% of tumors previously exposed to hormonal therapy.
Another large cluster, constituting 13% of these tumors, had mutations in a signaling pathway called MAPK. This pathway is an alternative route to growth that cancer cells can use to compensate for the loss of estrogen.
“MAPK mutations give the cancer cells a lifeline to survive the effect of hormonal therapy,” says Pedram Razavi, a physician-scientist at MSK and the paper’s first author. He notes that many of these mutations had not previously been linked to resistance in patients.
A third group, encompassing about 9% of cases, had mutations in genes that modify the function of the estrogen receptor. Known as transcriptional regulators, these too were mostly previously unknown to be involved in resistance.
The researchers found that these mutations were largely mutually exclusive. “That tells us that there are several possible ways that cancer cells can become resistant to hormonal therapy,” Dr. Razavi says.
Powerful Dataset
A key strength of the study was the integration of genetic data and clinical information from each patient’s medical history. This permitted the researchers to identify which mutations became more common in tumors over time, as women were treated with hormonal therapies.
“It’s not easy to make these kinds of clinical–genetic correlations,” Dr. Baselga says. “A tremendous amount of work, from several different teams at MSK, went into this analysis.”
Physician-scientist David Solit, Director of the Marie-Josée and Henry R. Kravis Center for Molecular Oncology (CMO), and computational biologist Barry Taylor, Associate Director of the CMO, were key collaborators on the project.
A limitation of the study is that it is a retrospective analysis based on correlations. Once the team identified these correlations, they went back to the lab and recreated the genetic changes in cancer cell lines maintained in the laboratory. With these experiments, they were able to show, for example, that MAPK mutations did indeed produce resistance to a class of hormonal drugs called selective estrogen receptor degraders. Furthermore, they were able to overcome this resistance by targeting some of the key steps in the MAPK pathway. These laboratory results help bolster the findings of the retrospective analysis and suggest specific strategies to combat resistance.
Impact on Treatment
The identification of MAPK pathway mutations as a cause of resistance to hormonal therapy reveals potential new drug targets. Several already existing US Food and Drug Administration–approved drugs target this pathway. These include HER2 inhibitors, EGFR inhibitors, ERK inhibitors, and MEK inhibitors.
The MSK team is eager to start clinical trials in people combining hormonal therapies with these targeted drugs to forestall — and perhaps prevent — the development of drug resistance.
Another upshot of the findings is that people who are known to have one of these mutations should probably not receive aromatase inhibitors as a treatment for their cancer, since the drugs will not be effective.
Unfortunately, genetic testing for these mutations is not routine and is not always covered by insurance, so this is not yet a part of care for many people with breast cancer.
Even if such tests were covered by insurance, it’s not practical to biopsy a person’s tumor or tumors at each visit to monitor the emergence of these mutations. A better solution, Dr. Razavi says, might be to use a liquid biopsy of a person’s blood to detect tumor-derived mutations in circulating cell-free DNA. A few such tests are available and several others are in development.
“We’re starting to do this type of testing now at MSK, and we hope that this will soon become standard of care,” he says.