Accurate transmission of the genetic information requires complete duplication of the chromosomal DNA each cell division cycle. However, the idea that replication forks would form at origins of DNA replication and proceed without impairment to copy the chromosomes has proven naive. It is now clear that replication forks stall frequently as a result of encounters between the replication machinery and template damage, slow-moving or paused transcription complexes, unrelieved positive superhelical tension, covalent protein-DNA complexes, and as a result of cellular stress responses. These stalled forks are a major source of genome instability.
The encounter of replisomes with damage in the leading- and lagging-strand templates have different consequences. As long as unwinding by the helicase is not impaired, damage in the lagging-strand template will not hamper replication fork progression in any significant way. This is because replication on that strand is already geared to constantly initiate new Okazaki fragments. Thus, a lagging-strand polymerase that does become stalled at a template lesion will be able to cycle forward rather soon to the next primer synthesized. Therefore the net effect of lagging-strand template damage is that gaps are left behind in the nascent lagging strand that have template damage directly opposite the 3¢-end of the gap (similar to the case in Fig. 1e, although the gap shown there is in the nascent leading strand).
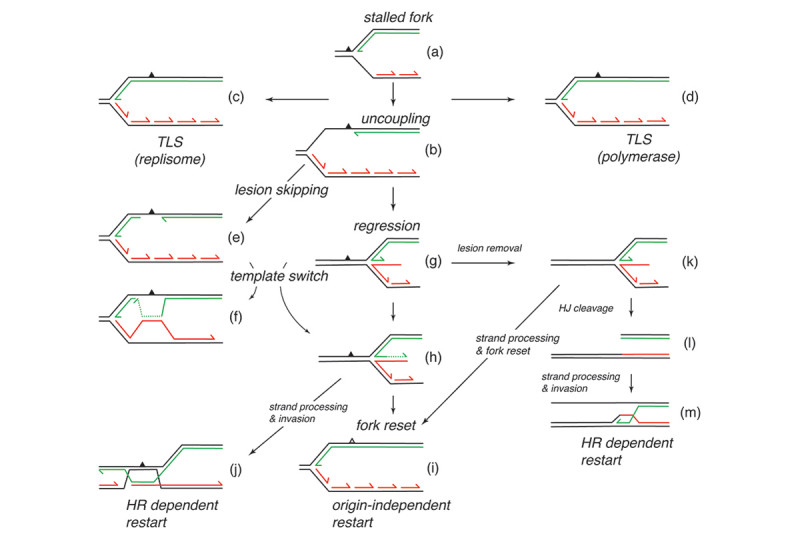
Damage in the leading-strand template is more problematic (Fig. 1). When a replisome first stalls at leading-strand template damage, the situation drawn in Fig. 1a obtains. At this juncture the nascent lagging strand has not yet caught up with the stalled leading strand. There are many different pathways that can be operative once the replisome has stalled (Fig. 1). We have used replication systems reconstituted with purified E. coli replication proteins and DNA templates carrying a single, site-specific cyclopyrimidine dimer (CPD) to analyze the events that occur after the replisome stalls. A major finding was that the template damage was only a transitory block to replication fork progression. Template unwinding and lagging-strand synthesis continued at a reduced rate downstream of the damage, exposing the leading-strand template in single-stranded form. A subsequent de novo priming event on the leading-strand template allows the stalled leading-strand polymerase to cycle forward to the new primer terminus and coordinated replication then continues downstream (Fig. 1a to 1b to 1e and Fig. 2). Thus, the replisome bypasses the template damage in a reaction termed “lesion skipping” leaving a gap behind. These studies indicated that the replisome was therefore inherently DNA damage-tolerant.
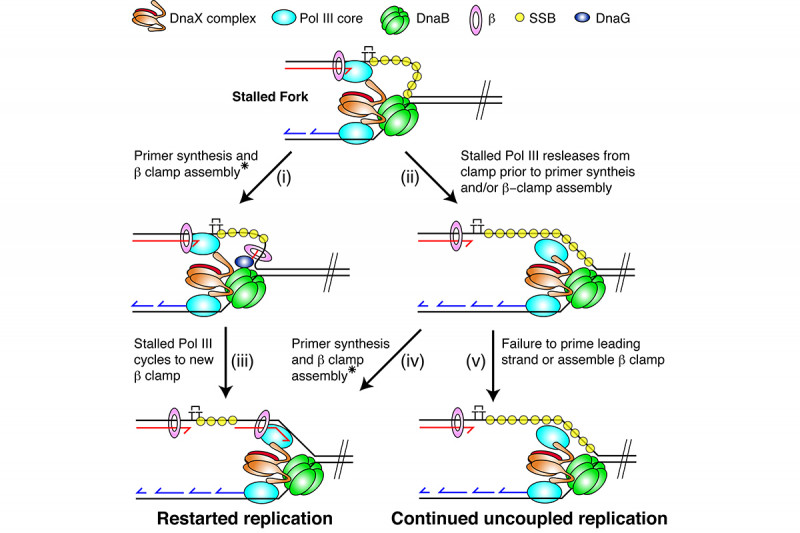
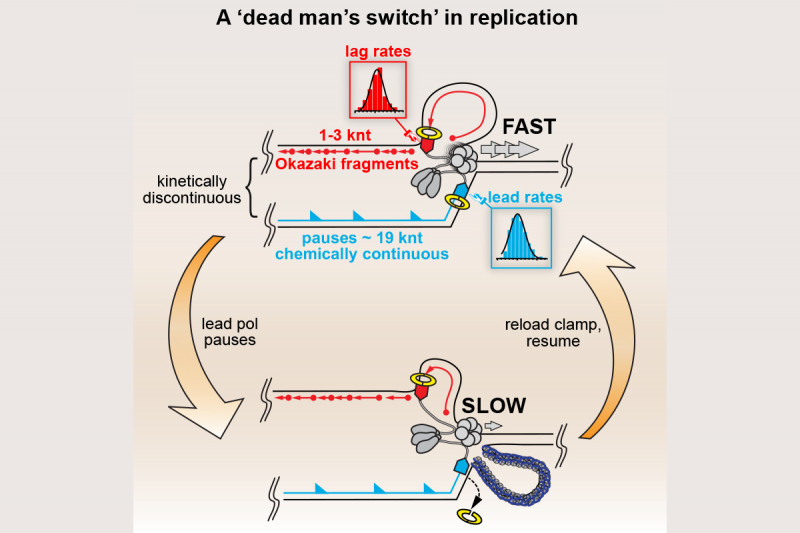
In collaboration with Steve Kowalczykowski’s group we subsequently demonstrated that the so-called polymerase uncoupling event (i.e., where, as described above, the leading-strand polymerase remains stalled while template unwinding and lagging-strand synthesis proceed downstream) that is thought to occur is actually a manifestation of an intrinsic lack of coordination between the leading- and lagging-strand polymerases. The two DNA polymerases at the replication fork do not communicate at all. Their behavior is completely independent and stochastic. They each sample from the same distribution of possible polymerization rates and they each exhibit pausing and switching of polymerization rates. Thus, at any given time the lagging-strand polymerase could be moving faster than the leading-strand polymerase and vice-versa. To prevent the accumulation of unreplicated gaps in the nascent DNA, when the leading-strand polymerase pauses the rate of the DNA helicase unwinding the parental template is reduced to about 15% of the normal rate. Once the leading-strand polymerase resumes DNA synthesis, it catches up to the helicase and rapid template unwinding and replication can continue (Fig. 3). We have termed this fail-safe mechanism a “Dead-man’s Switch.”
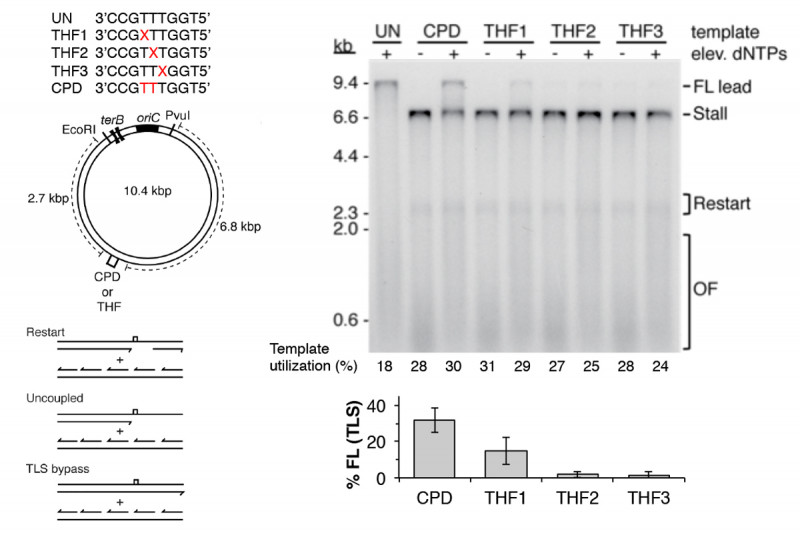
Another mechanism that contributes to DNA damage tolerance is direct bypass of template lesions by trans-lesion synthesis (TLS). In the past it was generally assumed that all TLS was accomplished by specialized DNA polymerases that could tolerate bulky lesions in the template at their active site and which switched with the stalled replicative polymerase (Fig. 1a to 1d). We showed that only DNA Polymerase IV (DinB) and not DNA Polymerase II, both of which are bypass polymerases, could associate with a stalled replisome to bypass a CPD. Recently we discovered another pathway of DNA damage tolerance, direct lesion bypass by the replicative polymerase in the replisome (Fig. 1a to 1c). We found that that the DNA Polymerase III holoenzyme, which is the replicative DNA polymerase in the replisome, (prepared from a strain deleted of the genes for all the TLS polymerases and with the proofreading exonuclease fully active) itself could bypass both a CPD and an abasic site analog (Fig. 4). TLS came at the expense of lesion-skipping replication restart. Thus, the stalled leading-strand polymerase was likely cycling between proofreading and polymerization at the 3¢-end of the nascent leading strand. Whereas polymerization across from the CPD will be slow compared to the normal chemical step, it was still faster than the amount of time required for progression of the DnaB helicase downstream and the synthesis of a new leading-strand primer to enable the lesion skip. Remarkably, bypass only occurred when the Pol III HE was integrated in the replisome. The enzyme itself could not bypass template damage in a primer-extension assay. These findings suggest that DNA damage at the replication fork can be replicated directly by the replisome without the need to activate error-prone pathways.
Current projects in the lab focus on determining the signal for induction of the SOS response (the bacterial DNA damage response) after replication forks stall and on investigating the consequences of collisions between the replisome and the transcription machinery before and after DNA insult.