My laboratory studies how mRNA elements that do not encode amino acids contribute to the regulation of biological complexity. We examine how 3′ untranslated regions (3′UTRs) regulate protein functions without affecting protein abundance. We also investigate the role of mRNAs as cytoplasmic organizers that enable subcellular compartmentalization required for the implementation of protein functions.

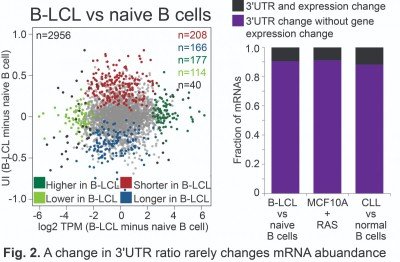
For many years, it was thought that the major function of 3′UTRs is the regulation of protein abundance (Mayr, 2018, CSH perspect biol). However, quantitative mapping of 3′UTR isoforms across tissues and metabolic conditions revealed that only a small fraction of significant 3′UTR ratio changes is associated with changes in mRNA abundance (Fig. 2) (Lianoglou, Genes Dev 2013). This raised the question of ‘What are 3′UTRs doing? (Mayr, CSH perspect biol 2018).
3′UTRs regulate protein functions

|
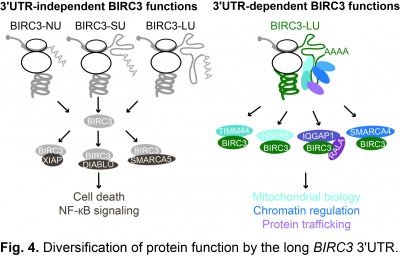
In 2015, we discovered that mRNAs do not only encode the amino acid sequence of proteins, but contain additional information on proteins. We found that 3′UTRs encode information for the protein interaction partners of newly translated proteins. Our studies revealed that 3′UTRs recruit protein interactors to the site of translation and the 3′UTR-dependent protein interactors control the subcellular localization and function of the newly translated proteins (Berkovits & Mayr, Nature 2015). For example, in the case of the plasma membrane protein CD47, we found that the long 3′UTR is required for efficient plasma membrane trafficking (Fig. 3). For the non membrane-bound E3 ubiquitin ligase BIRC3, we observed that 3′UTRs can diversify protein functions independently of protein localization (Fig. 4). In addition to the regulation of cell death, which is a 3′UTR-independent function of BIRC3, the protein encoded from the mRNA isoform with the long 3′UTR can also regulate B cell migration by controlling trafficking of CXCR4 (Lee & Mayr, Mol Cell 2019).
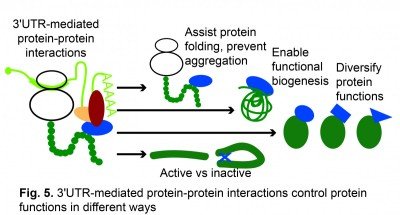
3′UTRs use co-translational protein complex assembly to control protein functions in different ways. Recently, we found that 3′UTRs are also able to control protein activity without affecting protein levels (Mitschka & Mayr, Kwon & Mayr, unpublished results) (Fig. 5).
mRNA-induced cytoplasmic organization through membraneless organelles
Our finding that 3′UTRs regulate diverse functions of proteins with identical amino acid sequence indicates that 3′UTRs contain genetic information for protein functions. We next investigated how information stored in 3′UTRs is transferred to proteins and focused on formation of the 3′UTR-dependent interaction between CD47 and SET. This led to the discovery of a membraneless organelle, called TIS granule that is formed through assembly of the RNA-binding protein TIS11B. TIS granules are constitutively expressed in all cell types and form a reticular meshwork that is intertwined with the endoplasmic reticulum (Fig. 6).

The current focus of the lab is to study how 3′UTRs control protein functions, to investigate the functions of TIS granules and to learn how mRNAs contribute to the cytoplasmic organization by identifying new membraneless organelles that are associated with membranes. Our goal is to identify biochemical reactions that are promoted or inhibited when taking place in membraneless organelles. Our working hypothesis is that the enriched mRNAs and their bound proteins generate subcellular microdomains with local properties that enable the formation of co-translational protein complexes and certain signaling reactions. These local reactions are required for proper protein and cellular function.