Gene regulation by microRNAs
A trove of small RNAs
A surprise of the early part of this century was the revelation that small RNAs preside over a previously hidden universe of negative gene regulation. Of particular interest was the discovery that eukaryotic cells process RNAs with double-stranded character into 21-22 nucleotide regulatory RNAs. In the RNA interference (RNAi) pathway, exogenous double-stranded RNA is converted into ~21 nt small interfering (siRNAs), which guide the cleavage and destruction of perfectly complementary transcripts. In a related pathway, endogenous transcripts containing hairpin structures are processed into ~22nt microRNAs (miRNAs) (Figure 1), which negatively regulate host transcripts by cleavage or by translational inhibition. Yet another pathway, termed the piRNA pathway, yields ~24-30 nt RNAs that play critical roles in genome defense, especially in the germline. Understanding the biochemical pathways and mechanistic strategies that produce and mediate the functions of these diverse small RNAs is of great interest, as is elucidating the specific roles of individual small RNA genes, particularly miRNAs and siRNAs.
Biogenesis of diverse small regulatory RNAs
Over the past decade, we have used computational and molecular approaches to annotate mi/si/piRNAs, and to elucidate mechanisms of how they are made and how they are sorted into appropriate Argonaute effector complexes. We have conducted this work in both Drosophila and mammalian systems. Some of the major themes of this work include the following (Figure 1). (1) There turn out to be many “non-canonical” strategies to generate miRNAs, involving unexpected cellular nucleases that normally process other types of transcripts. (2) Loading of different small RNAs into appropriate Argonaute proteins is critical for their appropriate function, and is therefore tightly regulated. (3) There are many steps at which the biogenesis of different small RNA substrates can be regulated by trans-acting factors.

Figure 1. Complexity of small RNA processing and loading pathways. This schematic includes the canonical Drosha-Dicer-Ago miRNA pathway, but includes also a variety of alternative miRNA biogenesis pathways, including Drosha-independent and Dicer-independent strategies. Moreover, the existence of endo-siRNA pathways in species such as Drosophila necessitates strict loading pathways to ensure that different Argonaute effector proteins associate with appropriate small RNAs.
Control of development, behavior and disease by microRNAs
Animal miRNAs generally exhibit only limited complementarity to mRNAs, typically matching to ~7 nt motifs near their 5’ ends (Figure 2). Thus, animal miRNAs tend to regulate large target cohorts. Paradoxically, many animal miRNA knockouts exhibit subtle phenotypes at best, and thus the biological imperative of miRNA regulation can be challenging to infer. This has led to notions that miRNAs are mostly for “fine-tuning” or “robustness”. Nevertheless, the founding miRNAs such as lin-4 and let-7 exhibit profound defects, and our studies have revealed a plethora of developmental and/or behavioral phenotypes in Drosophila miRNA knockouts. Moreover, we have often identified individual mRNA targets whose in vivo deregulation is critical for driving mutant phenotypes. For example, our studies identified transcription factors, RNA binding proteins, and signaling genes as critical miRNA targets during contexts such as development of peripheral sensory organs, wing, eye, and CNS, or during behavioral contexts such as egg-laying, rhythmic behavior, and locomotor activity (Figure 2).

Figure 2. Stringent in vivo target regulation by a neural miRNA. (Left) Shown are abdominal ventral nerve cords in wildtype and knockout of mir-iab-4/8 (∆mir) stained for Lk (to register segments) and for the miRNA target Hth. There is massive derepression in the miRNA knockout. (Right) Behavioral defect in ∆mir is largely attributable to derepression of Hth. In contrast to wt, ∆mir almost completely fails to lay eggs after mating. Heterozygosity for hth, achieved by two different alleles, strongly rescues this defect.
We have also reciprocal in vivo genomewide screens of miRNA activity, which can be uniquely performed in Drosophila using transgenic miRNA libraries that we constructed (Figure 3). We found using unbiased genetic screening that different miRNAs could elicit specific morphological phenotypes that often closely resemble deregulation of known developmental regulators or signaling pathways. In addition, we performed transgenic miRNA modifier screens to uncover miRNAs with unexpected genetic activities in cancer-relevant models. For example, in a screen in the Drosophila eye for modifiers of Notch ligand-dependent growth, we found a variety of miRNAs whose misexpression led to synergistic overgrowth and/or metastatic-like behavior (Figure 3). Dissection of these phenotypes can uncover new principles about in vivo miRNA function and genes that can influence growth and tissue patterning.
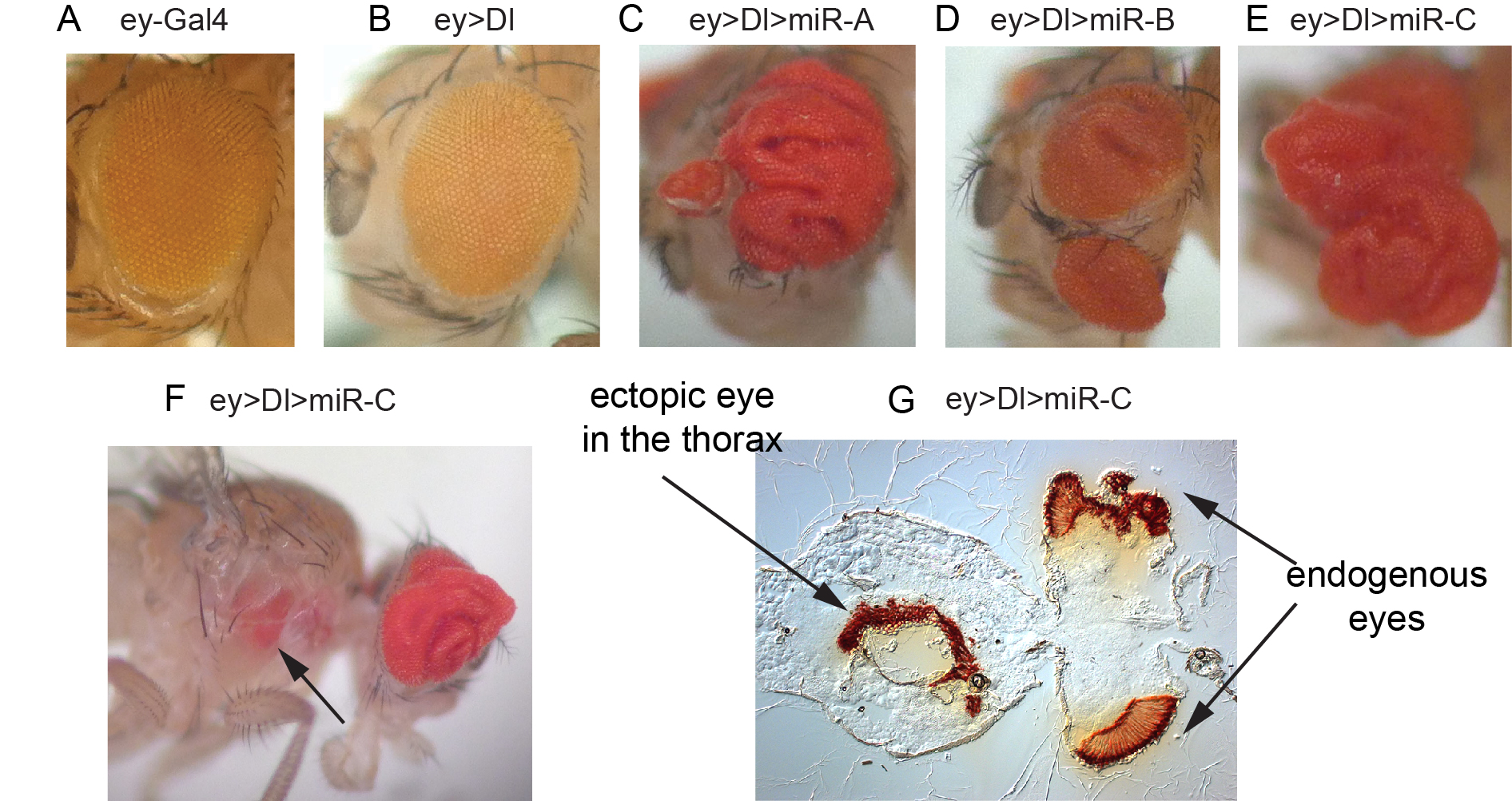
Figure 3. In vivo miRNA misexpression genetic screen. (A-E) Adult eyes that contain driver alone (A), express the Notch ligand Dl (B), or that coexpress Dl with different miRNAs that result in vast expansion of eye tissue (C-E). (F) Example of Dl-miRNA synergisms that yields migration of eye tissue to a distant location. (G) Sectioning confirm the presence of an ectopic eye in the thorax.