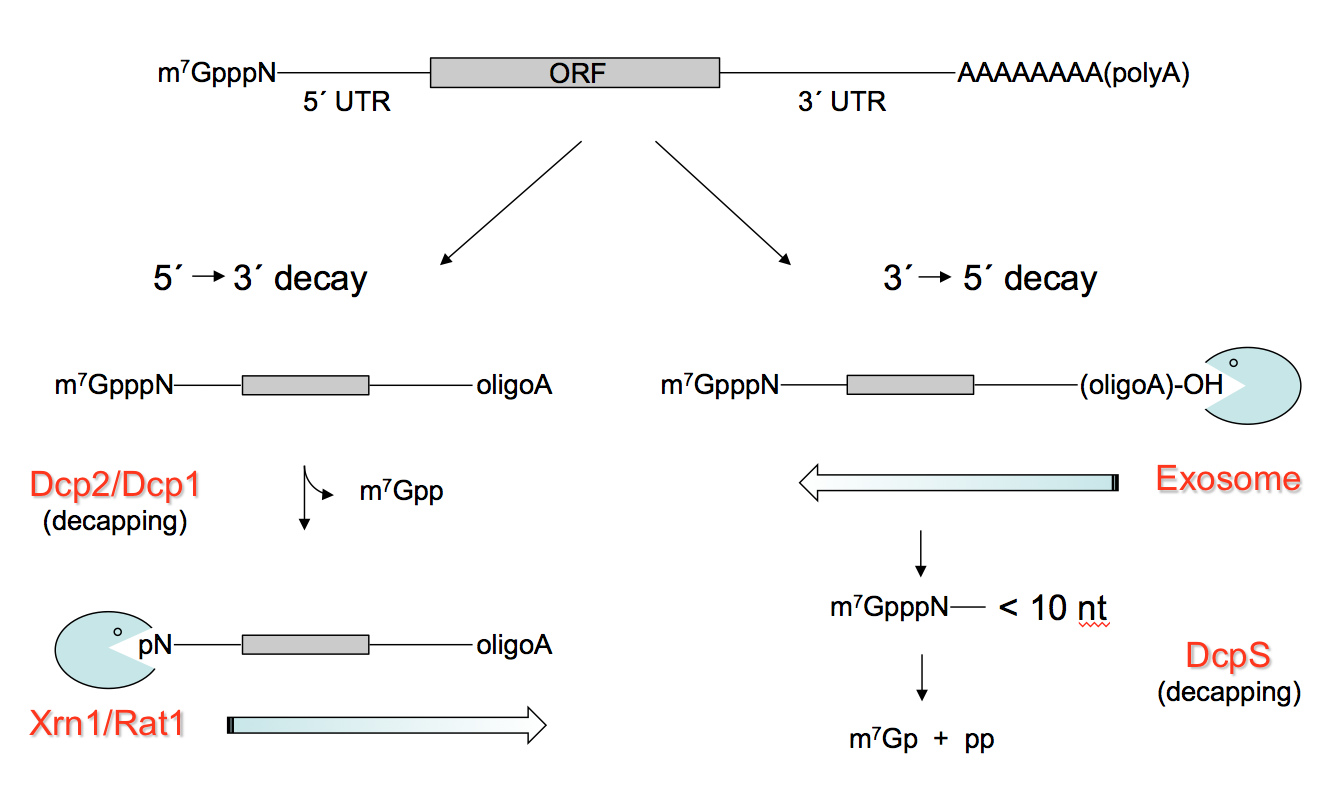
RNA abundance is regulated by balancing transcription and degradation in processes that control the temporal and spatial distribution of cellular RNA. In eukaryotic cells, mRNA decay is catalyzed by two major pathways, and both can be initiated by deadenylation of the polyadenylated (poly-A) tail. After decapping, 5’ to 3’ RNA degradation is accomplished by Xrn1, a 5’ to 3’ exoribonuclease. In the 3’ to 5’ pathway, RNA degradation is catalyzed by a multi-subunit 3’ to 5’ exoribonuclease complex, the RNA exosome. The exosome also participates in processing small nucleolar and small nuclear RNAs (snoRNAs, snRNAs) and ribosomal RNAs (rRNAs) and degradation of many non-coding RNAs including those that arise from bidirectional transcription or transcription from enhancers.
Composition of the eukaryotic exosome
The eukaryotic nuclear RNA exosome includes an essential nine-subunit non-catalytic core that associates with two ribonucleases, Dis3 and Rrp6 (EXOSC10 in human). The nine-subunit non-catalytic core shares structural similarities to bacterial 3’ to 5’ phosphorolytic exoribonucleases such as PNPase, including a prominent central channel that is large enough to accommodate single stranded RNA. While the eukaryotic exosome core is catalytically inert, its association with the catalytic subunits and additional cofactors that regulate the activities of Dis3 and Rrp6 on a variety of substrates.
Structures of RNA exosomes
We pioneered methods to reconstitute eukaryotic RNA exosomes from budding yeast and human. Early efforts determined the activities of yeast and human exosomes and the structure of the human eukaryotic nine-subunit exosome core.
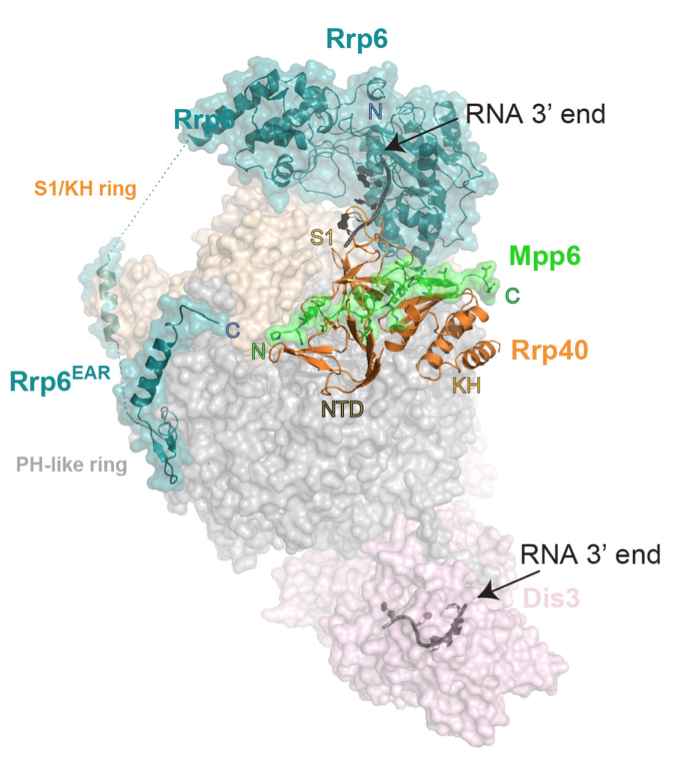
Our investigations of exosomes in budding yeast revealed that its central channel is essential, and that it functions to guide RNA substrates to the active sites of both Dis3 and Rrp6. We subsequently determined the structure of the yeast exosome core in complex with Rrp6 and polyA RNA, a high resolution structure of the eleven subunit nuclear exosome containing both Rrp6 and Dis3, and a twelve-subunit nuclear exosome bound to the Mpp6 cofactor. Collectively, this work illuminated RNA and subunit contacts to the exosome core that regulate its catalytic activities and recruit RNA helicases such as Mtr4 to promote helicase-dependent RNA decay.
Cyro-EM Structures of RNA exosome-helicase complexes
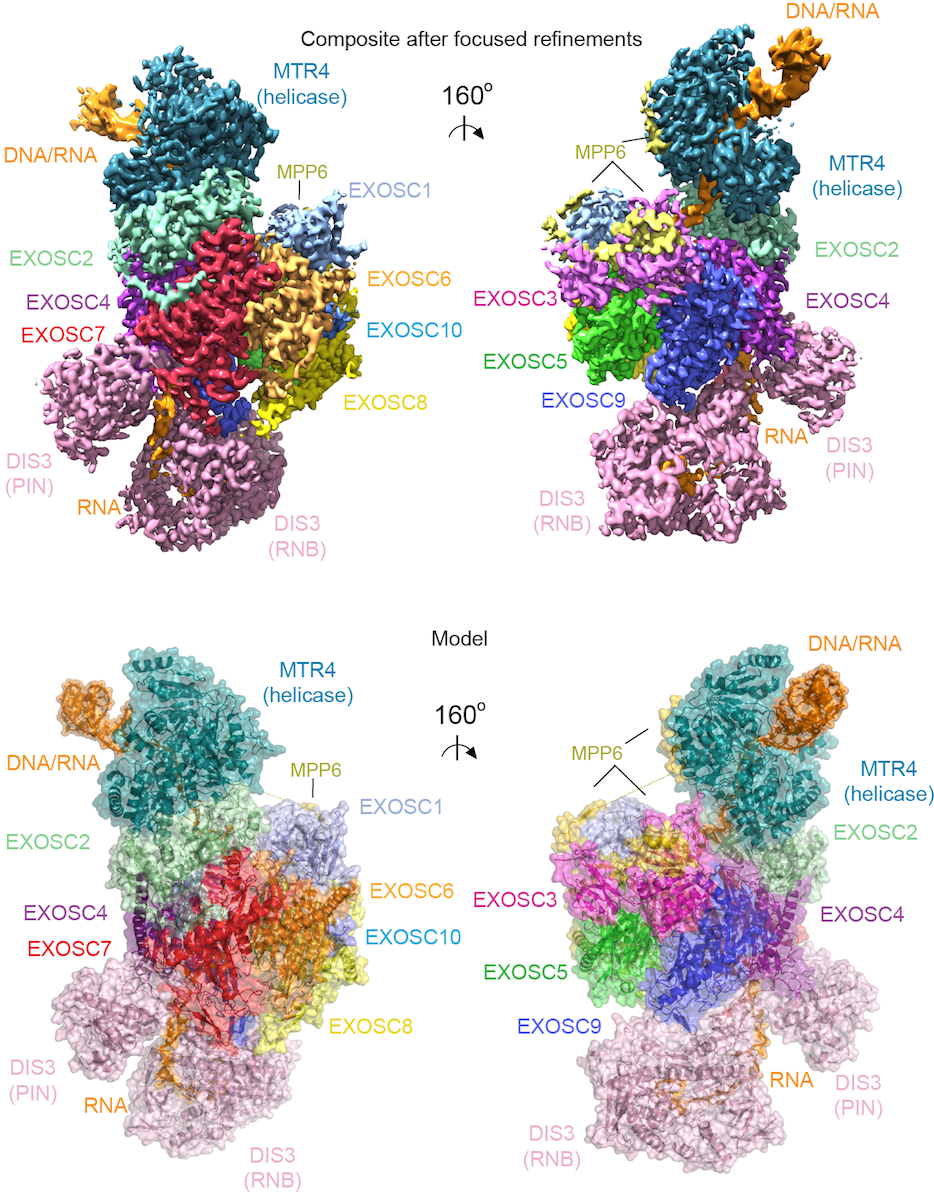
To understand how the 3’ to 5’ Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted fourteen-subunit Mtr4-containing RNA exosomes from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and human and show that they can all unwind structured substrates to promote degradation. We were able to load a human exosome with an optimized DNA-RNA chimera that stalls the MTR4 helicase during unwinding and determined its structure by single particle cryoelectron microscopy (cryo-EM) to an overall resolution of 3.45 Å. The structure reveals an RNA-engaged MTR4 helicase atop the non-catalytic core, with RNA captured in the exosome core central channel and DIS3 exoribonuclease active site. The MPP6 cofactor tethers the MTR4 helicase to the exosome through contacts to the RecA domains of MTR4. EXOSC10 (Rrp6 in yeast) remains bound to the core, but its catalytic module and cofactor C1D (Rrp47 in yeast) are displaced by RNA-engaged MTR4. Competition for the exosome core may ensure that RNA is committed to degradation by the exoribonuclease DIS3 when engaged by MTR4.