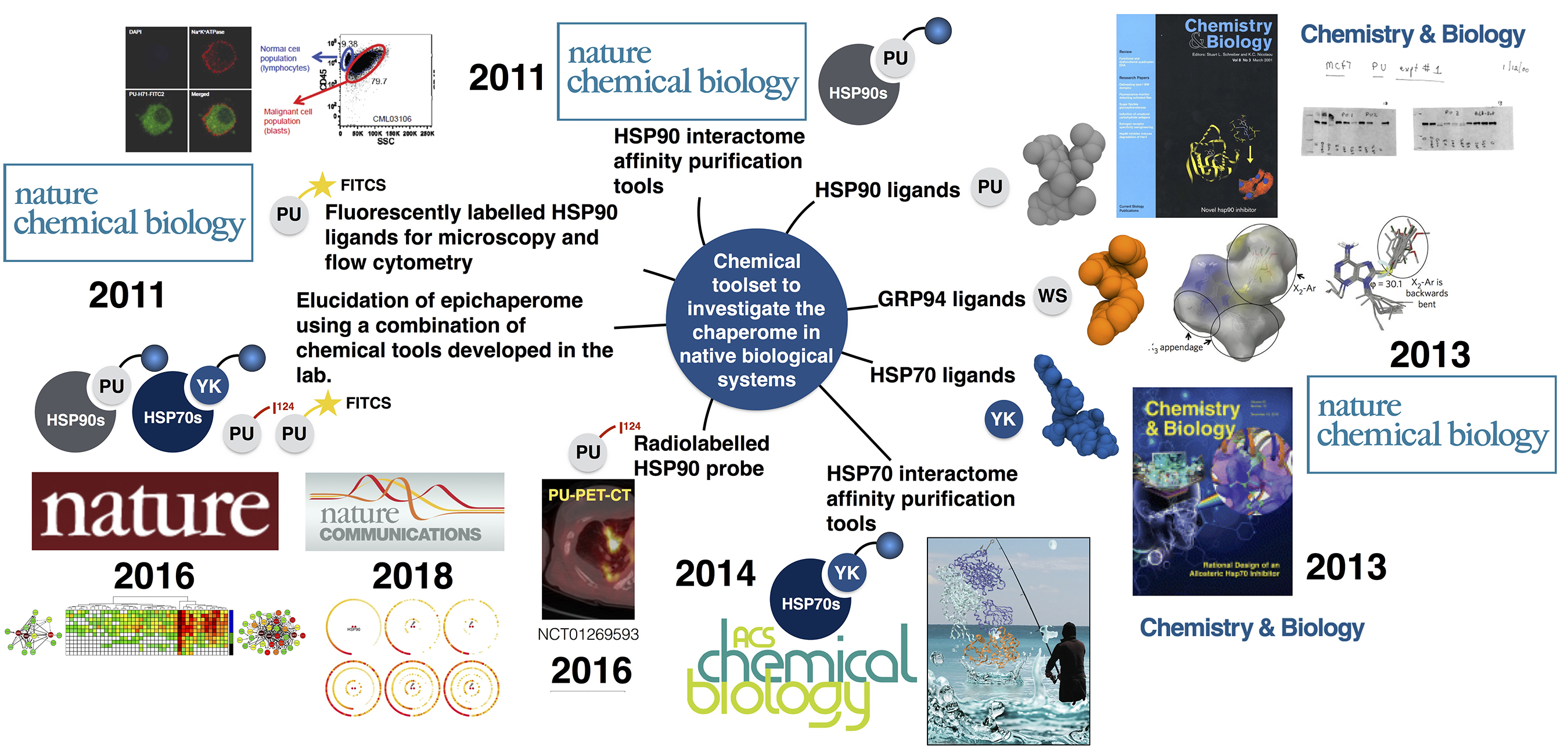
Continuously defining itself, the scope of chemical biology is broad and highly multidisciplinary. We use an approach to chemical biology wherein chemicals are “mutated” in order to obtain knowledge on a protein or a cellular state in a manner akin to how proteins are mutated in structural biology or cells are engineered in cell biology. These mutated chemicals are commonly referred to as probes or chemical probes, and we often also hear the term chemical toolbox or toolset, when not one but many of such probes are available to study and investigate a specific biological question. We use chemical probes to address the biology of proteins in the native cellular environment and to probe and manipulate a protein’s function in a controlled manner.
Chemical tools developed by my lab come in many flavors, including small molecules for cellular and organismal investigation of the CSC (i.e. ligands with in vitro and in vivo optimized properties such as the inhibitors PU-H71 for cancer and PU-AD for neurodegenerative diseases, the GRP94-directed PU-WS13 and derivatives for certain cancers, and the HSP70-directed YK5 and derivatives for cancers), appropriately labeled small molecules for investigations of the CSC by classical biological techniques (i.e. fluorescent probes for microscopy and flow cytometry such as PU-FITC; biotinylated or solid-support attached for large scale proteomic analyses by mass spectrometry such as PU-beads and YK-biotin; radiolabeled for use in imaging such as 124I-PU-H71 for cancer and 124I-PU-AD for Alzheimer’s disease). Each probe may have a specific or multifaceted role in addressing biology. Because these probes were derived by chemical intuition and not from screening, each pharmacophore represents a chemical entity never before made by man or nature – thus, we have also developed novel chemical methodology for their assembly and facile derivatization.
Our focus and deep interest on chemical biology has always transcended beyond basic biological investigation and into the realm of applying our understanding towards the treatment of human disease.
Chemical biology has played an important role in our understanding of the chaperome and proteome networks in disease, and has given answers to questions that were less amenable to being addressed with classical methods such as those routinely used in cell biology, genetics and biochemistry.
Together with medicinal chemistry, which provides the know-how to adjust these chemical tools to have the characteristics necessary for in vivo studies (i.e., favorable bioavailability and toxicology, proper in vivo target modulation), chemical biology promises to offer important insights into the function and regulation of the molecules that regulate life processes and, in turn, facilitate development of new therapeutics for the treatment and diagnosis of diseases.