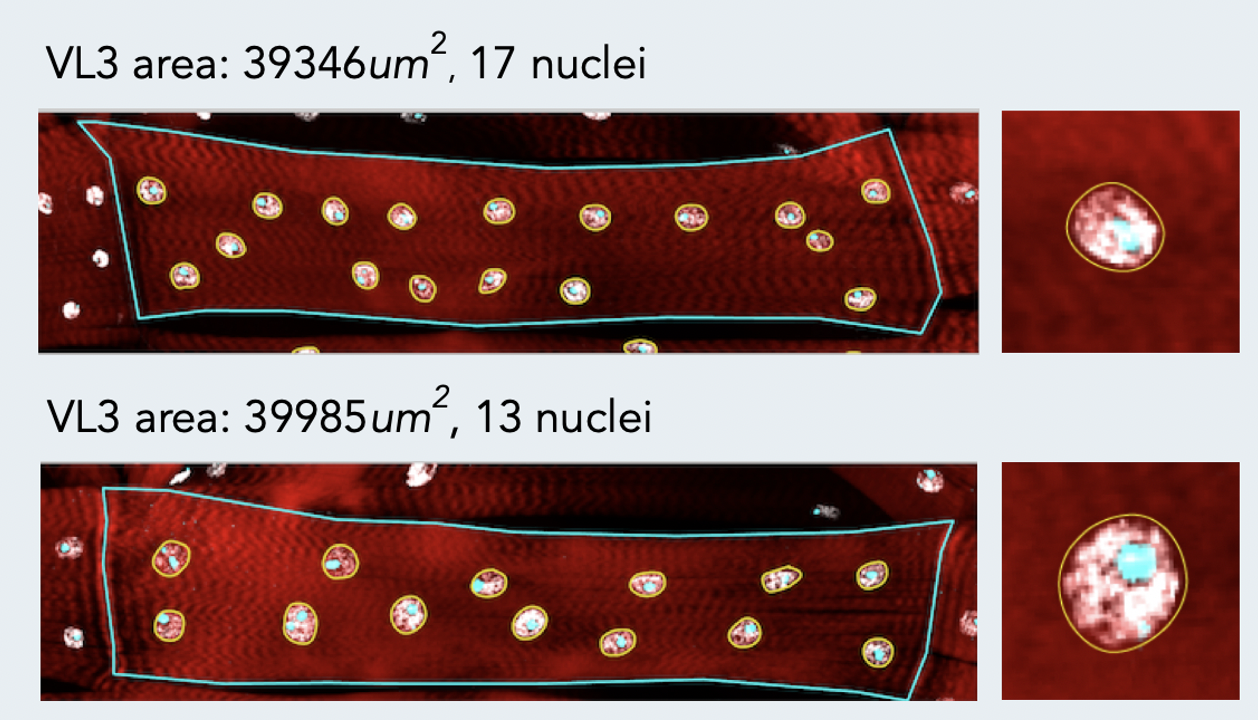
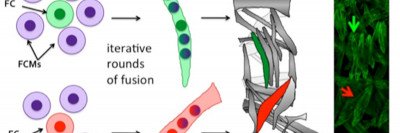
A key challenge in muscle biology is the generation and maintenance of a multinucleated muscle cell with a particular size, shape, and activity. To that end, we are actively defining those mechanisms that drive the formation of a muscle cell of a particular size. Recent work has focused on the contribution of nuclear number, size, position, and activity to muscle cell size and has revealed particular scaling relationships between these parameters and muscle cell size. We also find that nuclei within a single muscle cell have distinct scaling relationships depending on global, regional, and local inputs. These differences must be coordinated to generate a muscle of particular size. Current projects address the mechanisms by which nuclear identities arise, are maintained and are coordinated. We are also extending our studies to investigate scaling relationships and organization of other muscle cell organelles.