Control of cell fate and behavior by signaling and TFs
Notch signal transduction
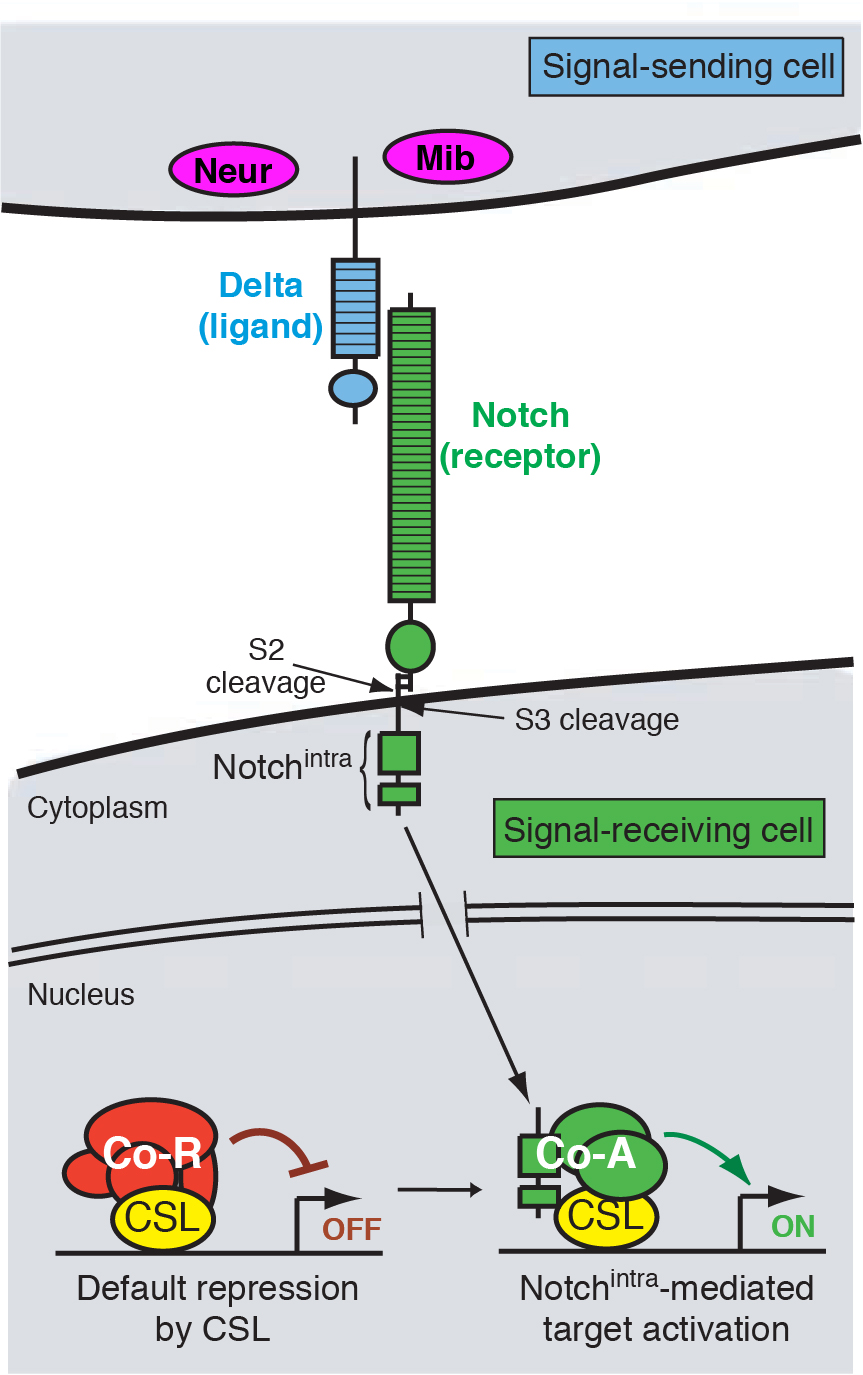
Coordinated development requires that cells communicate with each other. This is made possible by molecular mechanisms of cell-cell signaling, by which cells can influence each other’s fate and behavior. A cell signaling mechanism of fundamental importance to animal development is the Notch pathway (Figure 1). It may be safely said that Notch signaling is required, in some reasonably direct fashion, for the development of most tissues in all animal species. As such, misregulation of Notch signaling underlies a variety of human diseases and cancers.
Many ways to use a pathway
We focus on a few developmental settings that are paradigms of Notch-regulated events. One broad category is “inhibitory” signaling, whereby Notch prevents a cell from adopting a particular cell fate, usually through the repression of celltype-specific determinants. For example, Notch signaling singles out individual neural precursors from clusters of cells with neural potential. Loss of Notch signaling therefore causes excess neural differentiation. A second broad category is “inductive” signaling, whereby activation of Notch induces the expression of molecules with tissue-organizing activity. For instance, Notch signaling specifies the wing margin, a line of cells that organizes the patterning and outgrowth of the wing proper. Loss of Notch signaling here results in loss of wing tissue. We have utilized both inhibitory and inductive signaling events to study the roles of different Notch pathway factors, and how they regulate the activity of resident tissue and cell identity genes.
BEN proteins are novel CSL co-repressors in the Notch pathway
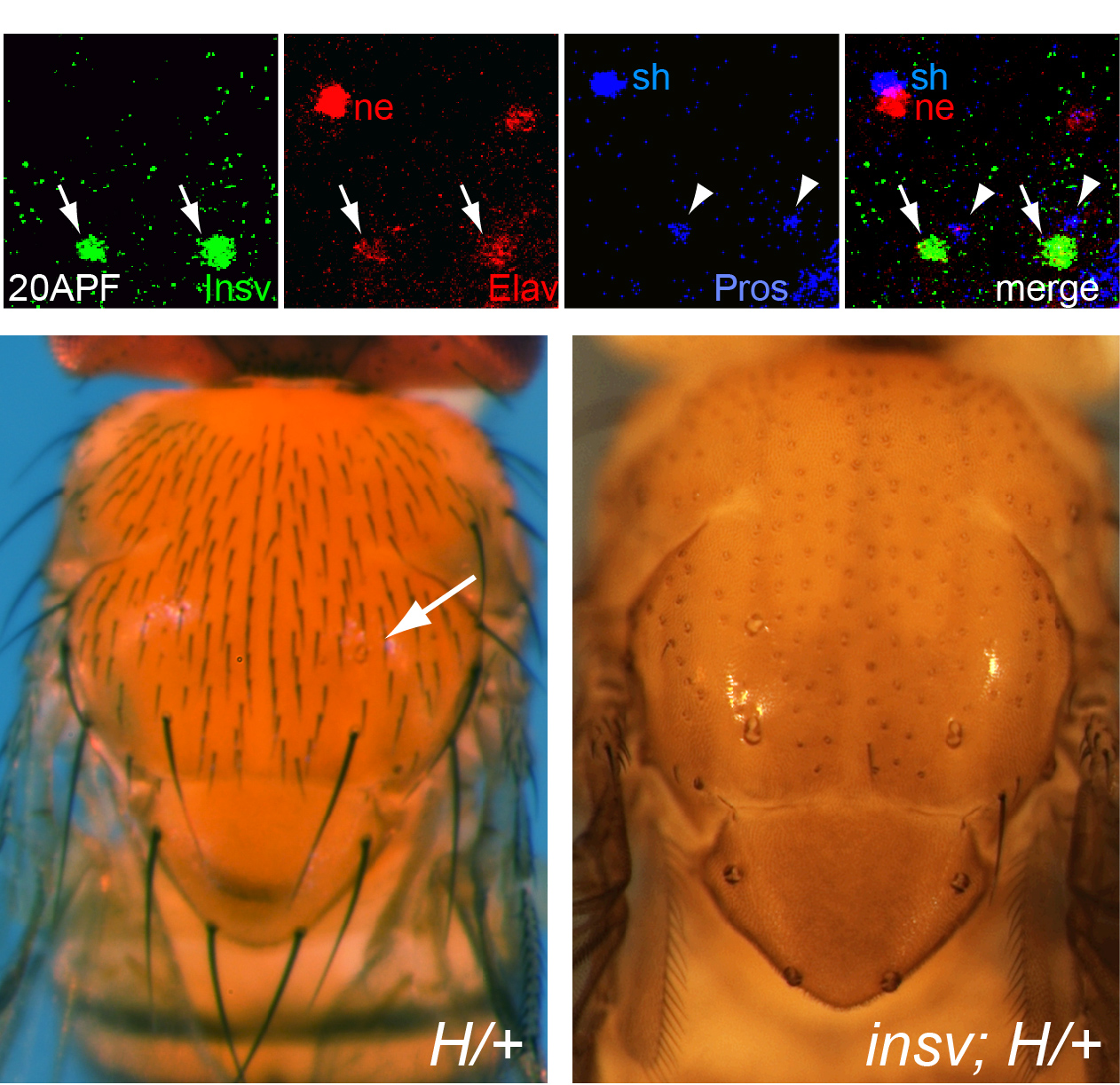
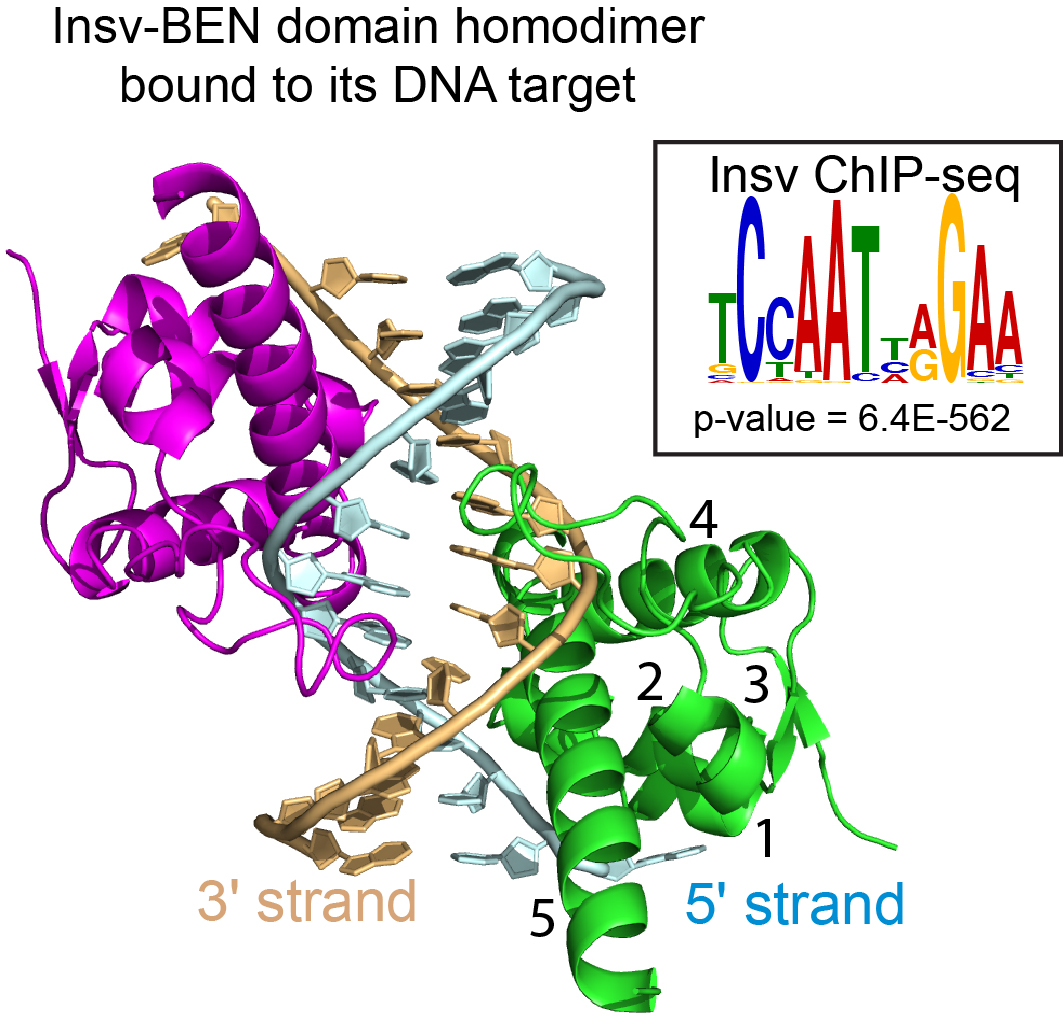
In the course of studying Drosophila novel factors that regulate neural development, we elucidated the molecular genetic function of Insensitive (Insv), an anonymous protein whose only annotated domain was of unknown function, the BEN domain. We showed that Insv is a nuclear factor that inhibits Notch signalling during multiple peripheral nervous system cell fate decisions. A direct role for Insv in transcriptional repression was indicated by binding of Insv to CSL transcription factor Su(H), and by strong chromatin immunoprecipitation of endogenous Insv to most E(spl)-C loci. Interestingly, we later found that the mammalian BEN domain factor BEND6 shares the functional attributes of Insv, even though they have only modest sequence similarity. BEND6 binds the mammalian CSL protein CBF1 and antagonizes Notch-dependent target activation. In vivo, BEND6 inhibited Notch-mediated self-renewal of neocortical neural stem cells and promoted neurogenesis. Thus, BEN domain factors represent a conserved type of Notch co-repressor in the nervous system.
The BEN domain reveals a new DNA binding domain