While we emphasize mechanism-based science, we have a long-standing interest in developing new technologies that increase the rate or resolution at which we can interrogate cancer-related processes. For example, we recently developed CRISPR-based methods to better model the genetic changes found in human tumors. One approach involves advances to base-editing technology that permit highly efficient construct delivery and editing in challenging models such as stem cells and organoids (1, 2). These advances enable systematic interrogation of allele variation in cancer. Another approach is a generalizable method, MACHETE (Molecular Alteration of Chromosomes with Engineered Tandem Elements), to readily reproduce cancer-associated deletions (3). We believe these methods will more accurately mimic the types of alterations observed in human cancer.
Because traditional mouse modeling is too slow to explore the range of genotypes that contribute to human cancer, we also focus on efforts to speed up production of genetically engineered mouse models of cancer. In one approach, we use “mosaic” mouse models that involve the genetic manipulation of stem cells or organoids in culture followed by orthotopic transplantation into syngeneic mice (4). In another, we combine multiple cancer-predisposing and inducible alleles in embryonic stem cell–based genetically engineered mouse models (ESC-GEMMs) to directly produce experimental cancer cohorts in F0 mice (5, 6). These systems also incorporate tet-regulated shRNA technology to enable inducible and reversible suppression of endogenous proteins at different disease stages (7, 8).
Finally, we are developing approaches to somatically engineer cancer-predisposing mutations in wild-type mice using tissue electroporation (EPO-GEMM). This approach allows the study of genotypic heterogeneity (by simply changing the electroporated plasmids) and mechanisms of tumor-host interactions (by changing the recipient strains). EPO-GEMMs of prostate, ovarian, and gastric cancers have been produced and validated and used to study cancer subtypes, tumor immune surveillance, and response to immune checkpoint blockade (Figure 1) (9-11). This relatively simple approach is at least ten times faster than germline methods.
Current research in the Lowe lab typically combines advanced modeling, genetics, and genomics approaches to interrogate cancer in a comprehensive way.
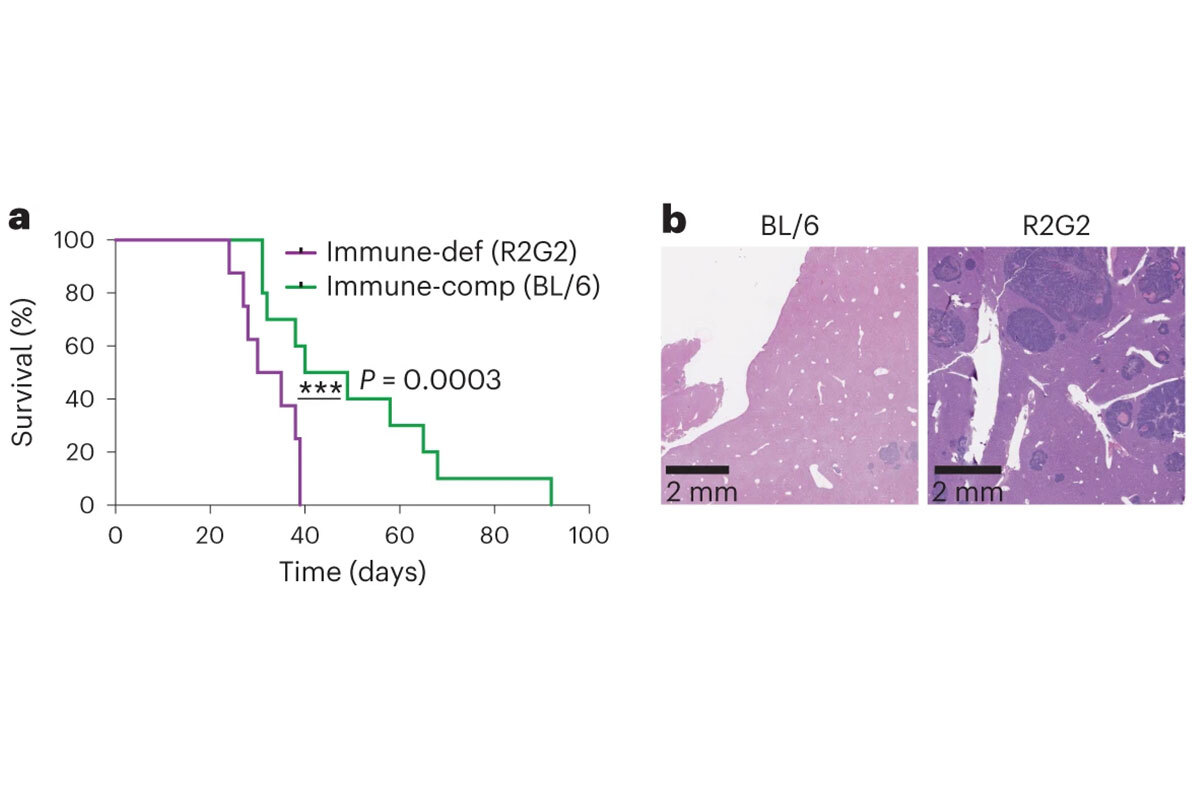
Figure 1. The ability to induce tumors via EPO-GEMMs allow rapid testing of the influence of genetic background on cancers. Here, in EPO-GEMM models of gastric cancer, immune-deficient R2G2 mice, compared with wild-type mice, showed more rapid death (a) and larger numbers of metastases (b), establishing a role for the immune system in limiting metastasis of these tumors (16).