Molecular Biology Services
Individual shRNAs
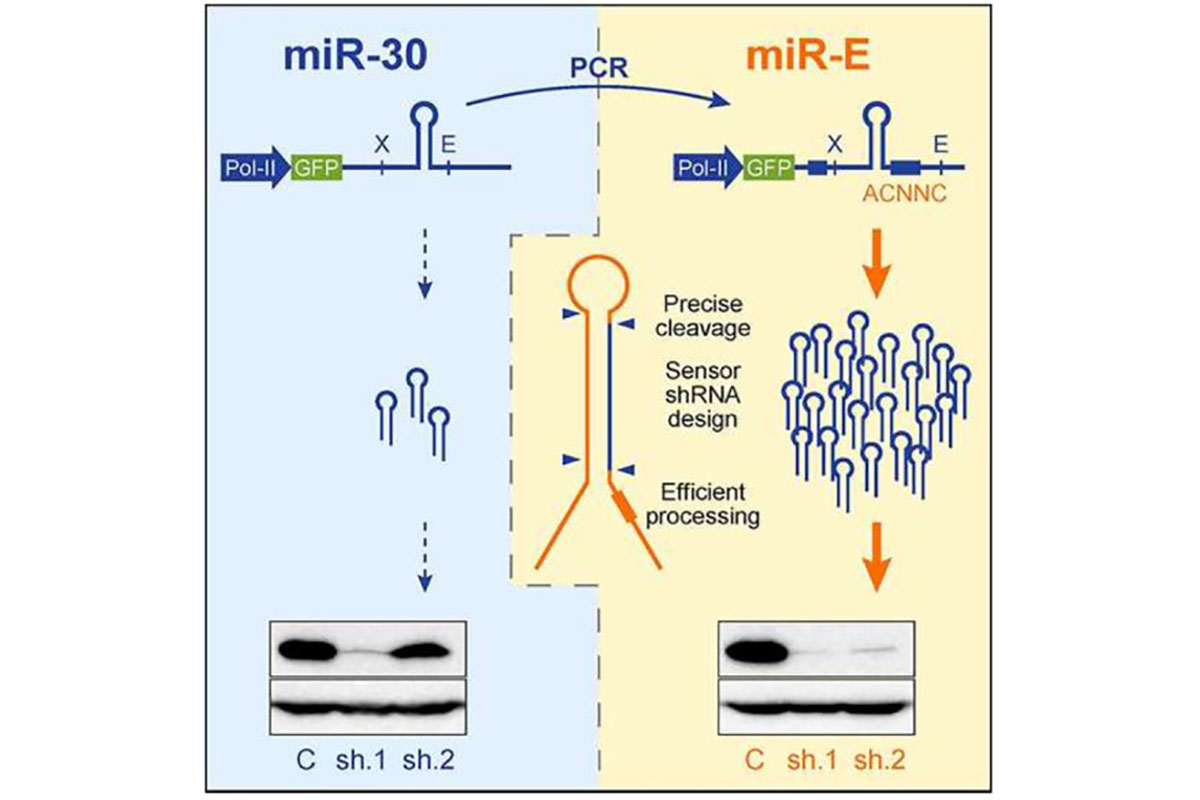
- We utilize the highly efficient mirE vector system for cloning your shRNAs
- shRNAs are designed using the SplashRNA algorithm. We embarked on collaborative work with fellow MSK researchers Scott Lowe and Cristina Leslie to develop a new sequential classification algorithm a sequential classifier (SplashRNA)1, to predict potent microRNA-based shRNAs. Combined with an optimized miR-E backbone2, >90% of high-scoring SplashRNA predictions trigger >85% protein knockdown.
-
Our miR-E vector collection includes a host of different selection characteristics to fit your experiments’ needs.
- Lentiviral or Retroviral Transduction
- Inducible or Constitutive Expression
- Fluorescence Reporters (GFP, dsRed, etc.)
- Antibiotic Selection Markers (Puromycin, Neomycin, Hygromycin, etc.)
- The mir-E vector panel is linked here so you may see our stock of vectors.
Multi-miR (Tandem) shRNA Cloning
- We offer tandem shRNA cloning in our mir-E vectors
- Tandem shRNAs are expressed off of the same promotor and allow for simultaneous knockdown of your genes of interest6
- Up to four shRNAs can be cloned and expressed off of a single promotor, while still providing high KD efficiency6
Individual sgRNAs
- We were involved with Andrea Ventura’s lab to qualify the GuideScan software for improved CRISPR guide RNA individual and libraries design. GuideScan produces high-density sets of gRNAs for single- and paired-gRNA genome-wide screens, that are more specific than those designed by existing tools3.
- We use lentiviral CRISPR-Cas9 vectors systems to clone your sgRNAs
- We offer both all-in-one (Cas9 inclusive) or multi-vector systems (your cell line expresses Cas9 from a separate vector)
-
Our vector selection includes a multitude of vectors with different characteristics
- Inducible or Constitutive Expression
- Fluorescence Reporters (GFP, dsRed, etc.)
- Antibiotic Selection Markers (Puromycin, Neomycin, Hygromycin, etc.)
Overexpression Cloning
- We also offer overexpression cloning
- We clone your coding region of interest into overexpression vectors
-
These vectors have multiple selection characteristics
- Fluorescence Reporters
- Antibiotic Selection Markers
Custom shRNA & sgRNA Pooled Libraries
- We offer custom designed, barcoded shRNA or sgRNA libraries
- Sufficient number of negative or possitive controls will be included in the design
- Cloning can be done into a wide choice of retro or lentiviral vectors
- Libraries are Maxiprepped and then are sent for QC using Next Generation Sequencing Platforms to ensure that all of your hairpins or guides are properly represented
“Off the Shelf” shRNA Pooled Libraries
| Library | Number of Genes | Subpools | Number of shRNA per gene | Control |
|---|---|---|---|---|
| shRNA Human Druggable Genome | 2,263 | 6 | 5 | Yes |
| shRNA Human Cancer Metabolome | 639 | 2 | 5 | Yes |
| shRNA Human DNA Damage Repair | 1,825 | 5 | 5 | Yes |
| shRNA Mouse DNA Damage Repair | 310 | 6 | 10 | No |
| shRNA Human Kinome | 526 | 3 | 5 | Yes |
| shRNA Human Epigenome | 565 | 2 | 5 | Yes |
| shRNA Human Phosphatase | 254 | 1 | 5 | Yes |
| shRNA Human Nuclear Hormone Receptor | 49 | 1 | 5 | Yes |
| shRNA Human Oncogene (1) | 62 | 1 | 6 | Yes |
| shRNA Human Tumor Suppressor (1) | 73 | 1 | 6 | Yes |
| shRNA Mouse Oncogene (1) | 62 | 1 | 6 | Yes |
| shRNA Mouse Tumor Suppressor | 73 | 1 | 6 | Yes |
| shRNA Mouse Phagosome-Proteosome | 1,385 | 10 | 6 | No |
| shRNA Mouse Drugged Genome | 446 | 3 | 5 | Yes |
| shRNA Mouse DNA Methylation | 495 | 5 | 5 |
Yes |
“Off the Shelf” sgRNA Pooled Libraries
| Library | Number of Genes | Subpools | Number of sgRNA per Gene | Control |
|---|---|---|---|---|
| sgRNA Human SAM Library | 23,430 | 1 | 3 | Yes |
| sgRNA Human Brunello Library | 19,114 | 1 | 4 | Yes |
| sgRNA Mouse Brie Library | 19,674 | 1 | 4 | Yes |
| sgRNA Human Epigenome | 565 | 1 | 4 | Yes |
| sgRNA Human Druggable Genome | 2,263 | 6 | 4 | Yes |
| sgRNA Human GeCKO Library V2 | 19,050 | 2 | 6 | Yes |
| sgRNA Mouse GeCKO Library V2 | 20,611 | 2 | 6 | Yes |
| sgRNA Human GeCKO Library V1 | 18,080 | 1 | 2 - 4 | Yes |
| Human Chromatin | 1,624 | 2 | 6 | Yes |
| Human Transcription Factor | 1,918 | 2 | 6 | Yes |
| Human Metabolism | 2,976 | 1 | 6 | Yes |
| Human DNA Damage Repair (DDR) | 989 | 1 | 6 | Yes |
| Human Saturn v2.0 | 3,136 | 5 | 6 | Yes |
| Mouse Saturn v2.0 | 3,136 | 5 | 6 | Yes |
“Off the Shelf” Barcoded Libraries
| Library | Number of Barcodes | Subpools |
|---|---|---|
| Celltag | 19973 | 3 |
| LARRY | 245,979 | 1 |
| Watermelon | 5,000,000 | 1 |
CRISPRi/CRISPRa Cloning
- We have a stock of CRISPRactivation and CRISPRinhibition vectors
- CRISPRa/i utilizes catalytically inactive Cas9 (dCas9) and an sgRNA complementary to either transcriptional start sites (TSS) or promotor regions to repress/activate gene expression5
- The dCas9 can also have transcriptional activation or repression cassettes (VP64, VRP, Mxi1, etc.) added to increase activation or repression efficiency4
Genetic Perturbation Screening Services
Given the technical challenges associated with making these complex measurements, it is imperative to have resident Core expertise embedded at the institution, to assist all MSK investigators. Essential features of a genetic perturbation screening shRNA/sgRNA based include:
gDNA Extraction, NGS Sample Prep, & Analysis
- gDNA extraction from cells pelleted by centrifugation
- PCR amplification of region of interest and NGS
- Deconvolution and analysis of Hiseq quantification to find enriched and/or depleted shRNA(s)/sgRNA(s) in the screenings based upon unique identifying barcodes. The analysis is performed using R and in particular edgeR. edgeR is designed for finding changes between two or more groups. It is a Bioconductor software package for examining differential expression of replicated count data and the methodology can be used even with the most minimal levels of replication.
Screening Performance
- We offer shRNA and sgRNA individual and pooled library screening
- Assay development including viral transduction optimization.
- Screening execution, genomic DNA extraction, and PCR for next generation sequencing.
Cell Line Generation Services
We generate, through RNAi or CRISPR-Cas9 gene editing, knockdown (KD), knockout (KO) and knock-in (KI) isogenic cell lines respectively for genes of interest (GOI) or long non-coding regions (Lnc), with isolation and verification of single clones for modeling human disease. We have experience working with a wide variety of cell backgrounds ranging from easy-to-transfect cell lines to hard-to-transfect cell lines.
-
mir-E-based KD Cell Line Generation
- We generate stable shRNA expressing cell line using the mir-30 technology. The gene knockdown temporarily stops or decreases the expression of one or more targeted genes, constitutive or inducible. If the cells survive a knockdown event, they can recover and eventually begin to express the gene as before. It is usually the best option for essential genes or if you want to mimic the effect of a drug, significantly lowering the production of a protein, yet keeping the mRNA transcription intact
-
CRISPR-based KO/KI Cell Lines
- Every editing project is unique, based on the modification desired and the cell background; therefore, we provide consultation services to evaluate the technical feasibility of editing in your desired cell line and to design the best strategy to achieve the goal. We design guides and repair templates, define the best backbone to use (all in one vector versus two vectors system), optimize the delivery strategy (infection, transfection or electroporation), the cell culture conditions and, and we validate per your specifications by CRISPR-Seq and Sanger Sequence to help ensure you receive the edited cell line you desire.
-
CRISPR-Seq
- We offer CRISPR KO/KI validation through Next Generation Sequencing
- The region of interest is PCR amplified and then sent for NGS
- Each amplicon gets 50,000 individual reads
High Content and High Throughput siRNA and Chemical Compound Screening
The mission of the High-Content and High-Throughput Screening Branch of the Gene Editing and Screening Core is to provide investigators access to the latest screening libraries and technologies. Core staff scientists are available to assist investigators in experimental and scientific design of their HCS/HTS experiments to ensure the correct protocols, reagents, and readouts are employed to best address the scientific questions of the investigator. We provide training and assisted support for many of our technologies from image acquisition to analysis. The HTS/HCS staff work closely with investigators with the goal of delivering high quality, reproducible, and rigorous data.
Arrayed Library Screening
- Assay development and semi-automated screening in 96/384-well plate density format.
- Cell viability assays and biochemical assays with EnVision multimode plate reader (PerkinElmer)
- High content cell imaging assay with IN Cell Analyzer 6000 laser-based confocal imaging platform (GE Healthcare) and Columbus image analysis system (PerkinElmer)
- High throughput live cell microscopy with phase contrast and green/red fluorescence on our IncuCyte S3 (Sartorius)
| Bioactives & FDA Drugs | # of Compounds | # of Plates (384) |
|---|---|---|
| Selleck Bioactive Library | 2,880 | 9 |
| Selleck FDA Approved Library | 1,280 | 4 |
| LOPAC 1280 | 1,280 | 4 |
| Selleck Epigenetics compound Library | 699 | 4 |
| Prestwick FDA Approved Chemical Library | 1,280 | 4 |
| MicroSource Spectrum Library | 2,560 | 8 |
| MedChemExpress Metabolism Library | 1,299 | 4 |
| MedChemExpress FDA-Approved Drug Library | 2,377 | 8 |
| Subtotal | 13,655 | 45 |
| Focused Set | # of Compounds | # of Plates (384) |
|---|---|---|
| Chembridge KinaSet Library | 3,200 | 10 |
| Chembridge KinaCore Library | 2,000 | 7 |
| GSK Published Kinase Inhibitor Set Library | 676 | 3 |
| Roche Kinase Set for MSK | 197 | 1 |
| MCE Drug Repurposing Compound Library | 3,418 | 11 |
| MCE Ferroptosis Compound Library | 526 | 2 |
| Enamine Phenotypic Screening Library | 5,760 | 18 |
| MedChemExpress Natural Product Library | 2,755 | 9 |
| Subtotal | 18,532 |
61 |
| Diversity Library | # of Compounds | # of Plates (384) |
|---|---|---|
| Chembridge CombiSet Library | 10,240 | 32 |
| Chembridge DiverSet Library | 10,240 | 32 |
| Chembridge PremiumSet Library | 10,240 | 32 |
| Enamine Discovery Diversity Set | 10,240 | 32 |
| Life Chemicals Diversity Set | 30,080 | 94 |
| Subtotal | 71,040 | 222 |
Imaging Services
INCell Analyzer6000
- We offer rental of our INCell Analyzer 6000 for you to image your 96 well or 384 well plates
- This system is capable of imaging fixed or live cells at 4X, 10X, 20X, and 40X
- Channels: FITC, CY5, DAPI, dsRED, and Bright Field
- Z-Stack images for organoids and other spheroid structures
- Reporter signals, antibody staining signals
- Lipid droplet/lipidTOX detection

Puncta identification
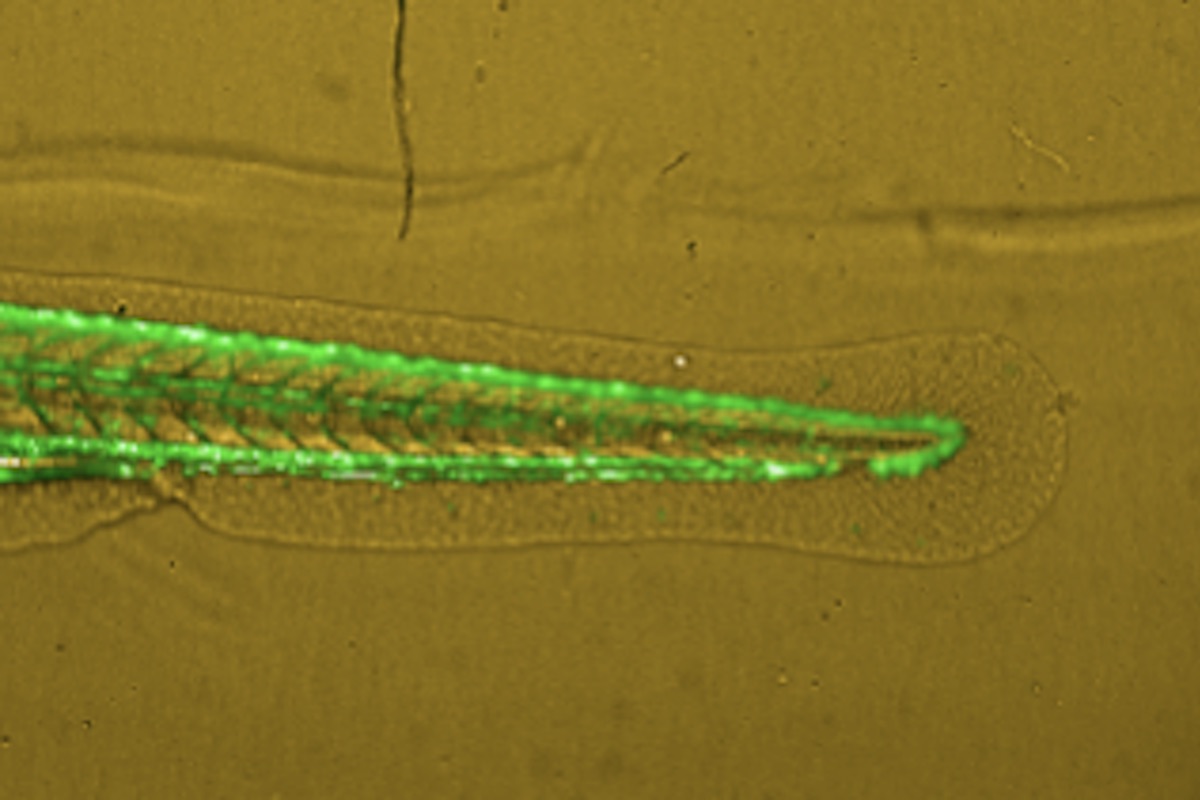
Zebrafish Based Cancer Migration Assay
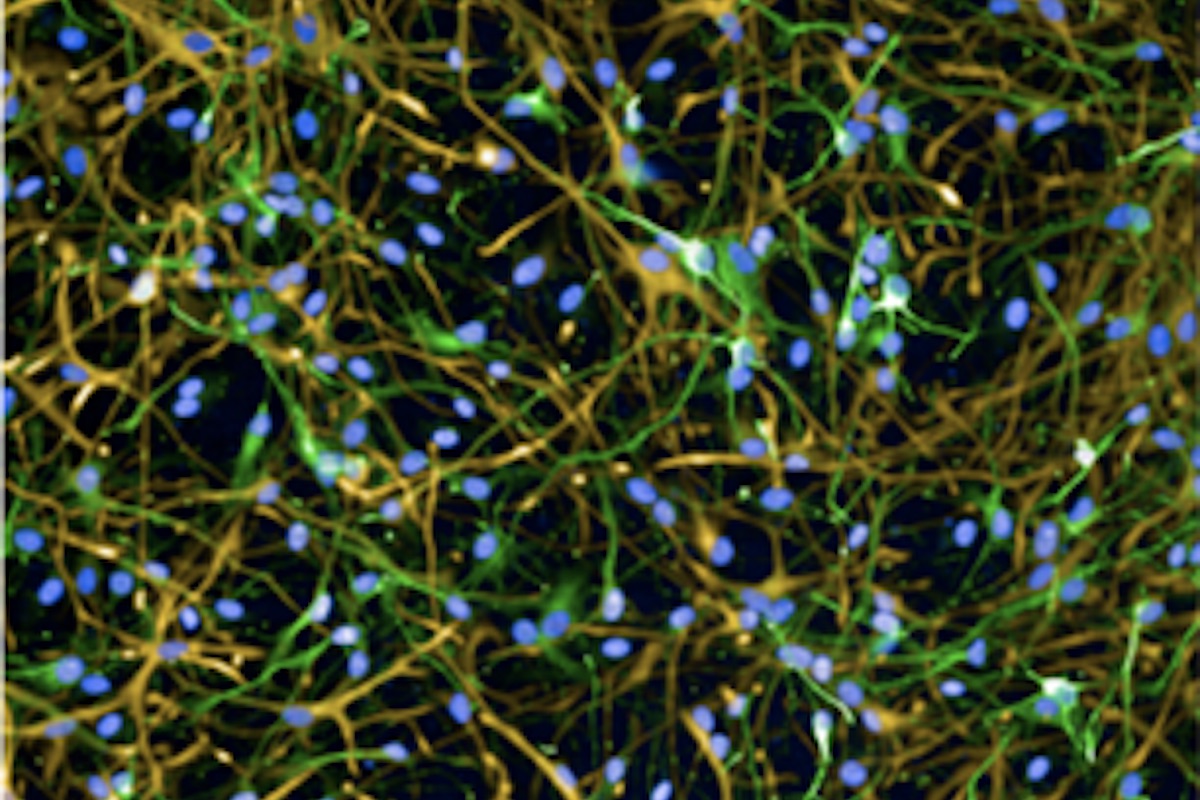
Astrocyte-Neuron co-culture Quantification Assay
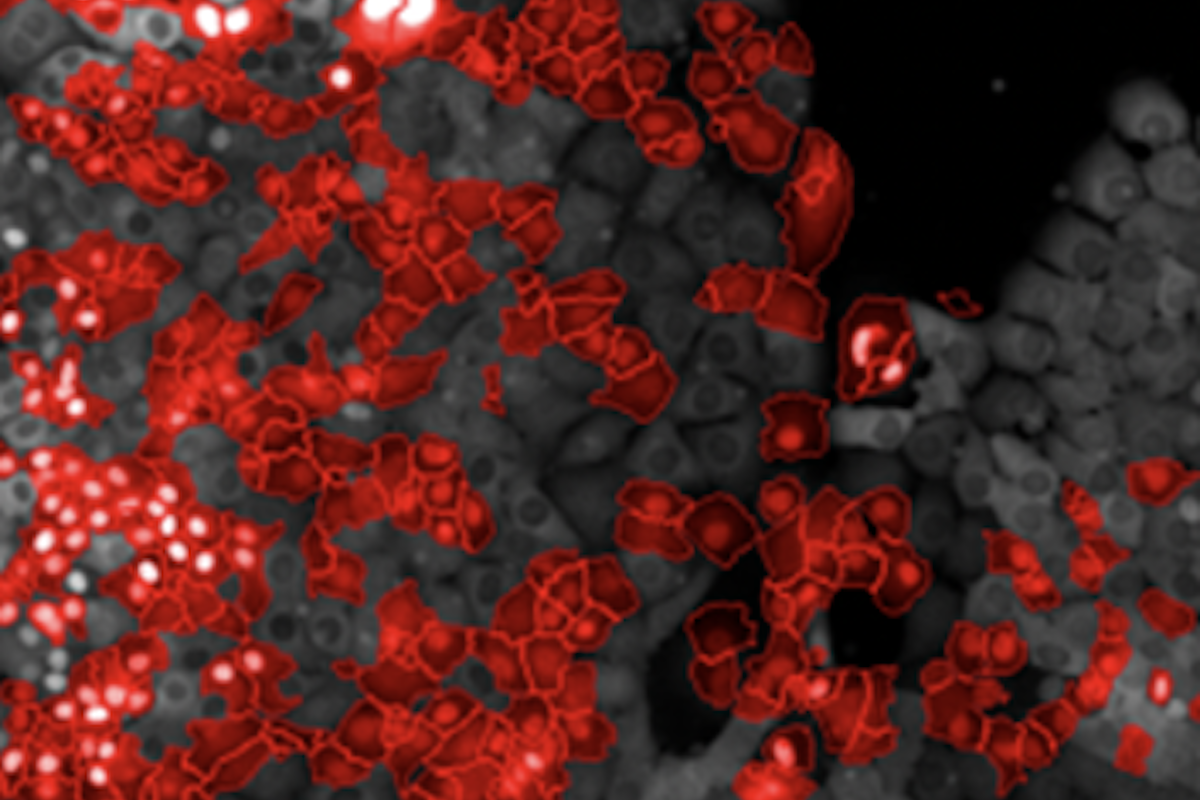
Nuclear Translocation Dectection
Academic Drug Discovery Consortium
- Co-hosted by Core Director Ralph Garippa, the Academic Drug Discovery Consortium is a professional society dedicated to the building of academic drug discovery labs and the companies, universities and government agencies that support and partner these centers
-
The ADDC focuses on a wide range of therapeutic strategies
- Small Molecules, Biologics, Vaccines, Antibodies, Gene Therapies, Cell Therapies
-
Activities spanning the drug discovery continum
- Biobanks, Biomarkers, Drug Delivery Systems, Toxicology, Animal Models and Facilities, Manufacturing, Diagnostic Tools, Clinical Trials
-
Mission
- Establish an interactive network of academic drug discovery scientists and centers (currently ~150 centers worldwide)
- Exchange expertise related to drug discovery programs, technologies, faculty engagement, intellectual property, regulatory agencies, industry, and other public-private partnerships
- Provide a platform for partnering and collaboration •between consortium members and the life science industry including pharma, biotech, CRO’s and other service providers
- Advocate for funding of academic drug discovery