
At Memorial Sloan Kettering Cancer Center, we have now analyzed over 65,000 samples in our prospective cohort with MSK-IMPACT™, our next-generation sequencing (NGS) platform, and are adding to that number every day. This is up 8000 from January this year, despite Covid-19.
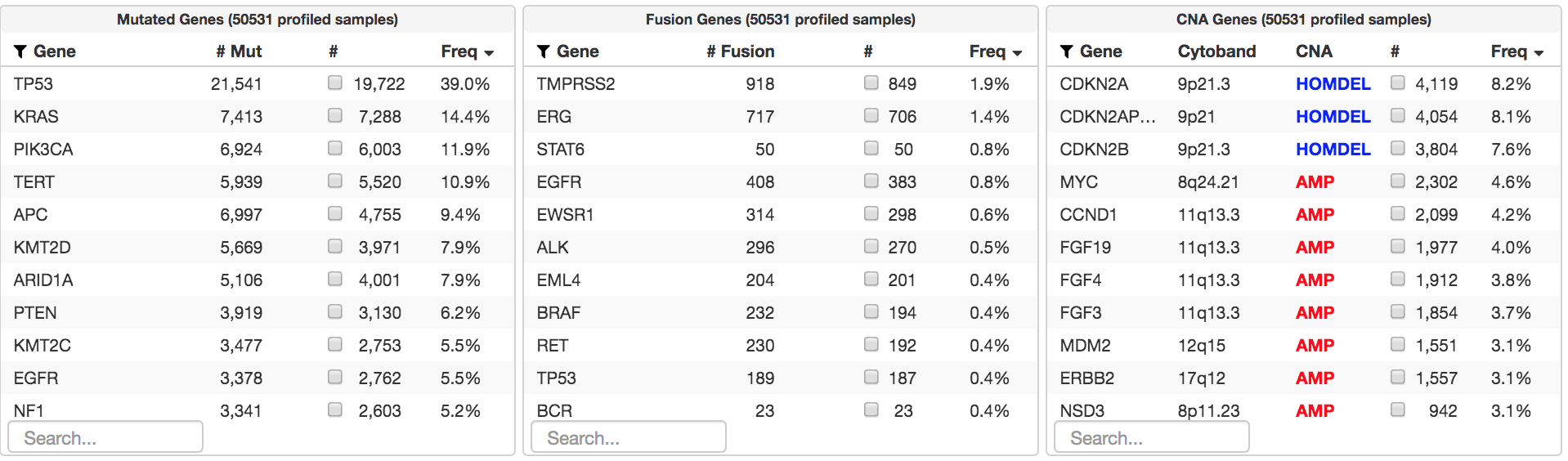
Tumor NGS should be considered for all patients with advanced cancer. It has historically been viewed as a companion diagnostic test for a subset of patients, specifically those who are candidates for systemic targeted therapies. This understates its clinical value. Tumor NGS is also critical to:
- ensuring the correct cancer type diagnosis, which impacts decisions regarding surgery, radiotherapy, and cytotoxic chemotherapy
- understanding a patient’s risk of recurrence and cancer-specific mortality
- assessing for heritable germline mutations
NGS is not overhyped. Some pundits view tumor NGS, and precision oncology more generally, as overhyped. In our experience, the results of tumor NGS directly dictate treatment in about 40 percent of patients with advanced cancer.
Why don’t more patients benefit from NGS? The main reason is that we do not have effective targeted therapies yet for many of the most common alterations identified in our MSK series to date, such as TP53, TERT, APC, and NF1. Increasing the fraction of patients who benefit will require the development of new drugs. Broader sequencing assays will have a more modest impact. For this reason, the development of new targeted therapies is the focus of many of the research laboratories and clinicians at MSK.
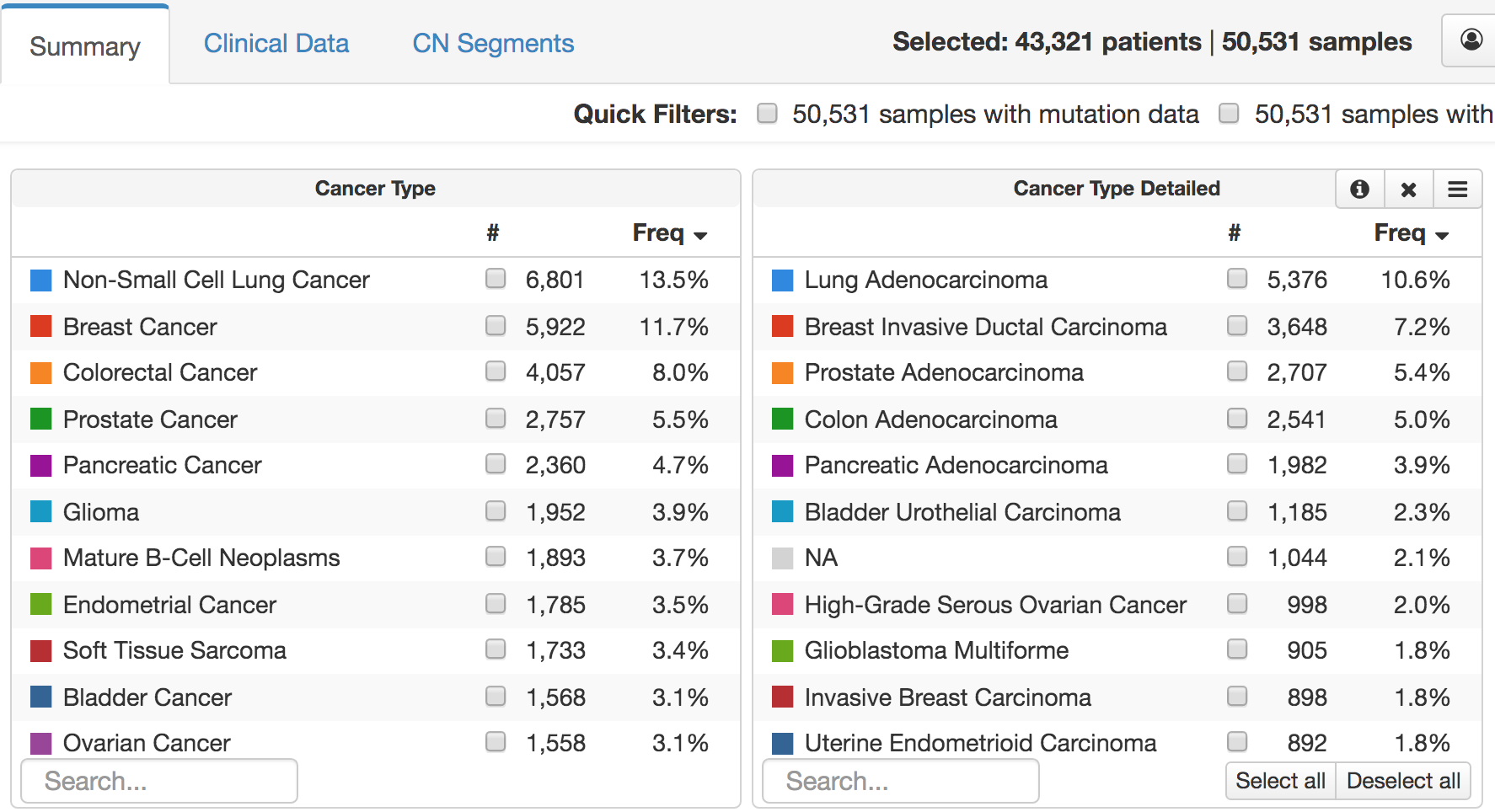
How much sequencing is enough for an individual patient? There are three general outcomes with tumor NGS:
- Identification of a compelling targetable mitogenic driver.
- Identification of a nontargetable driver sufficient to transform cells
- No identification of a mitogenic driver. In our experience, it is primarily this group that benefits from more profiling.
Note that in rare instances, the identification of druggable fusions missed by targeted NGS can still be impactful. In the future, identification of co-alterations in patients with a validated targetable driver may also prompt rational combinations of targeted therapies.
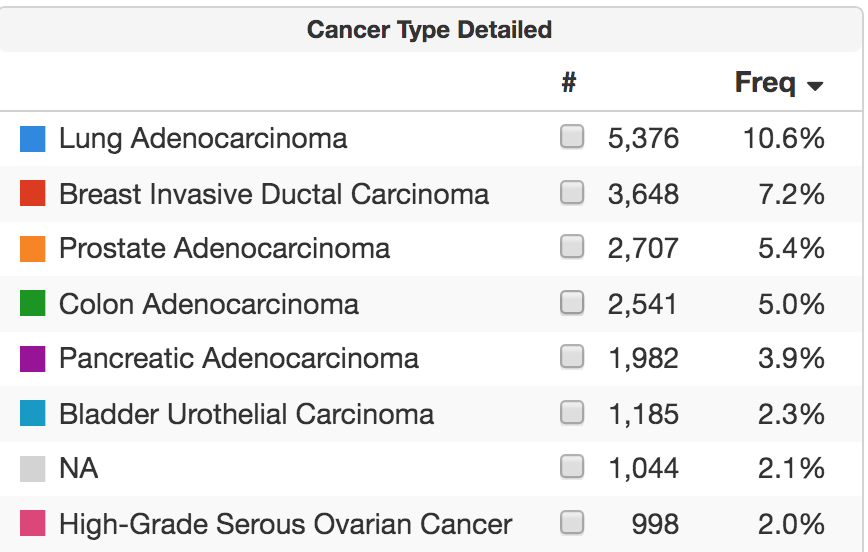
One hope for broader NGS is that it could identify mutational signatures that are predictive of drug sensitivity. This approach is now validated for microsatellite instability, found in 1,026 patients, 2.1 percent of our cohort to date. Whether whole-genome sequencing can identify homologous recombination deficiency or other signatures of value remains unclear.
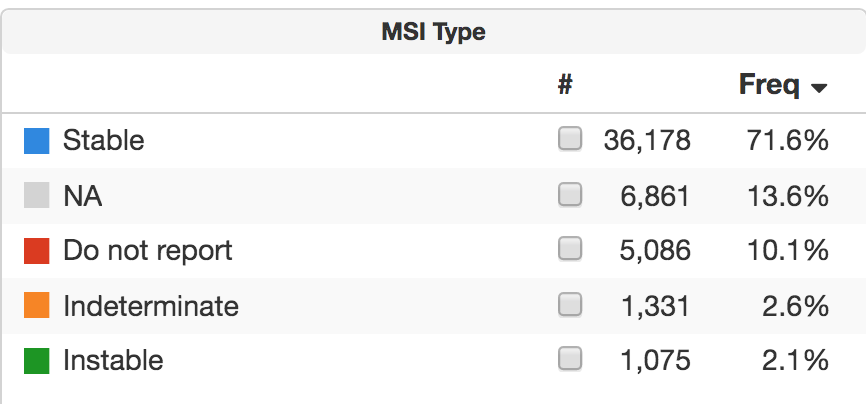
Why is “not available” listed as the cancer type for so many patients? The majority have suspected hematologic cancer, and the test was performed as an integral part of the diagnostic workup. This use of tumor NGS is increasingly common in solid tumor patients.
Yes, paired tumor-normal sequencing is the optimal approach. Why are we still debating this point? It seems increasingly clear that concurrent analysis of a “normal” sample is preferred to ensure that mutations are not germline variants or somatic mutations derived from clonal hematopoiesis.
Where does circulating free DNA (cfDNA) fit in? CfDNA has some distinct benefits over tumor NGS. Blood samples are easier to obtain and have a shorter turnaround time. Repeat samples are also easier to collect and may reflect tumor-to-tumor heterogeneity better. CfDNA does have some drawbacks. We must sequence at a much deeper level, which means a higher sequencing cost per gene and, typically, analyzing fewer genes. There is a possibility of false negative results due to low tumor DNA shedding. Sensitivity varies by alteration type, and there are more missense mutations than deletions. Finally, clonal hematopoiesis may confound results.
Given these pros and cons, it is likely that tumor and plasma NGS will prove to be complementary, not competitive, approaches. Clinicians will need to learn how to identify cases where initial cfDNA may have missed critical alterations due to a low tumor-derived plasma DNA fraction. At MSK, our liquid biopsy test, called MSK-ACCESS, captures 129 genes selected from MSK-IMPACT for high-sensitivity, noninvasive cancer genomic profiling and disease monitoring.
So where do we go from here? A critical next step is the ongoing collection of clinical treatment and outcome data for the tens of thousands of people with cancer who have participated in our MSK prospective sequencing initiative. We need to understand better why patients derive variable benefit from current therapies. This understanding will accelerate the development of more-effective treatments for patients who are not cured by existing approaches and to develop less toxic methods for those who are.
To facilitate this effort, MSK is a founding member of the American Association for Cancer Research’s Project GENIE (Genomics Evidence Neoplasia Information Exchange). This initiative is pooling genomic and clinical data from institutions worldwide, to understand better how specific genomic profiles impact the outcomes of people with cancer.
Together with my colleagues in the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, the Molecular Diagnostics Service, and Human Oncology and Pathogenesis Program at MSK, I hope we can fully realize the translational potential of tumor and plasma-based NGS over the next few years.
Dr. Solit has received honoraria and had a consulting or advisory role with Loxo Oncology, Lilly Oncology, Pfizer, Illumina, Vividion Therapeutics, and QED Therapeutics.

