
The High-Performance Computing (HPC) group maintains a wide range of computational resources to fit your needs. All are Linux compute clusters, each attached to large storage platforms to support intensive data research.
Each cluster is configured to meet the needs of its main users, with various operating CPUs and GPUs. Currently, Lilac is the primary cluster designated for new HPC research requests. Requests for alternate clusters are reviewed on a case-by-case basis depending on the research requirements and cluster availability.
- For access to HPC systems, visit the Getting Started page. To request an account, click here.
- The technical specifications for each cluster can be found here: https://rts.mskcc.org/available-computing-system
- To learn more about HPC services visit: https://rts.mskcc.org/high-performance-computing
- To request HPC services visit: https://rts.mskcc.org/service
Research HPC
The MSK and Sloan Kettering Institute research community has access to three HPC systems. Each has distinct differences that provide solutions to a unique set of computational requirements. Most users will operate on Lilac.
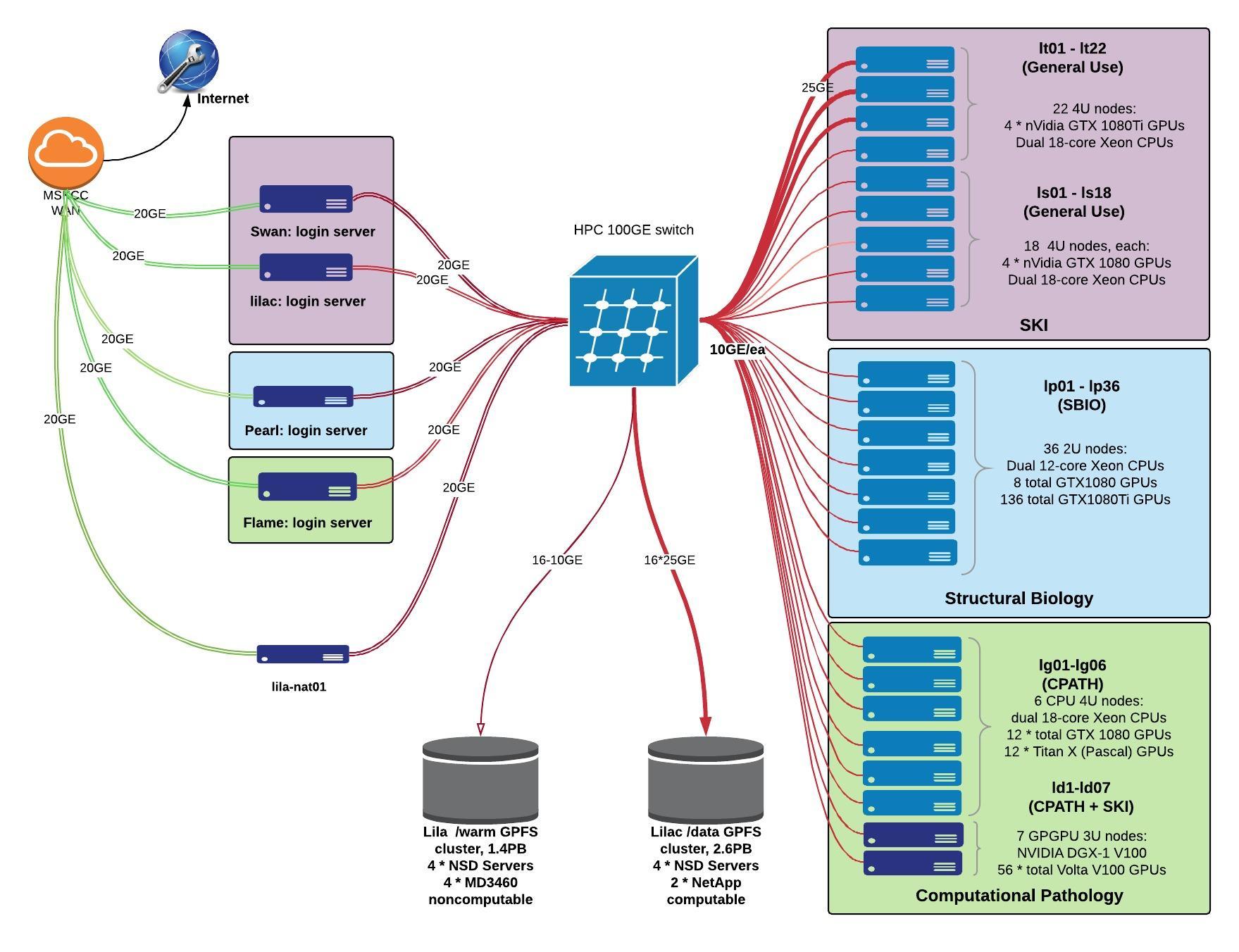
The Lilac HPC cluster runs an LSF queueing system, which supports a variety of job submission techniques.
Compute
- Primary login host: lilac.mskcc.org
- 91 compute nodes with GPUs. (GPUs on nodes vary): name quantity model CPU cores RAM GPUs NVMe net ld01-07 7 NVidia DGX-1 2*Xeon E5-2698 v4 40 512 8*Tesla V100 – 10 lg01-06 6 Exxact TXR231-1000R 2*Xeon E5-2699 v3 36 512 4*GeForce GTX TitanX – 10 lp01-35 35 Exxact TXR231-1000R 2*Xeon E5-2680 v3 24 512 4*GeForce GTX 1080Ti – 10 ls01-18 18 Supermicro SYS-7048GR-TR 2*Xeon E5-2697 v4 36 512 4*GeForce GTX 1080 2T 10 lt01-08 8 Supermicro SYS-7048GR-TR 2*Xeon E5-2697 v4 36 512 4*GeForce GTX 1080Ti 2T 10 lt09-22 14 Supermicro SYS-7048GR-TR 2*Xeon E5-2697 v4 36 512 4*GeForce GTX 1080Ti 2T 25 lv01 1 Supermicro SYS-7048GR-TR 2*Xeon E5-2697 v4 36 512 4*Tesla V100 2T 25
Storage
- /data (compute): 2.6PB, GPFS v5.1, 4 * NSD Servers, 4 * NetApp E5760, connected at 16*25gbps
- /warm (non-compute): 1.5PB, GPFS v5.1, 4 * NSD Servers, 4 * Dell MD3460, connected at 4*10gbps
Network
Arista 7328x 100gbps switch
Bioinformatics HPC
High-Performance Genomics Computing Systems Access
Computing resources for investigators in the Computational Biology Center and other SKI investigators who need access to significant Linux compute and storage resources specifically in association with the Center for Molecular Oncology or the Bioinformatics Core. These systems support processing of sequence data generated by IGO.
Three computational clusters support Bioinformatics operations. These systems are known as Lux, Luna, and Juno. Lux primarily supports processing of raw genomic data. Juno is currently the main computational system used within the Bioinformatics Core and by members of the Center of Molecular Oncology. For members of these labs; these systems provide access to shared research data, applications, and packages to support bioinformatics analysis pipelines.
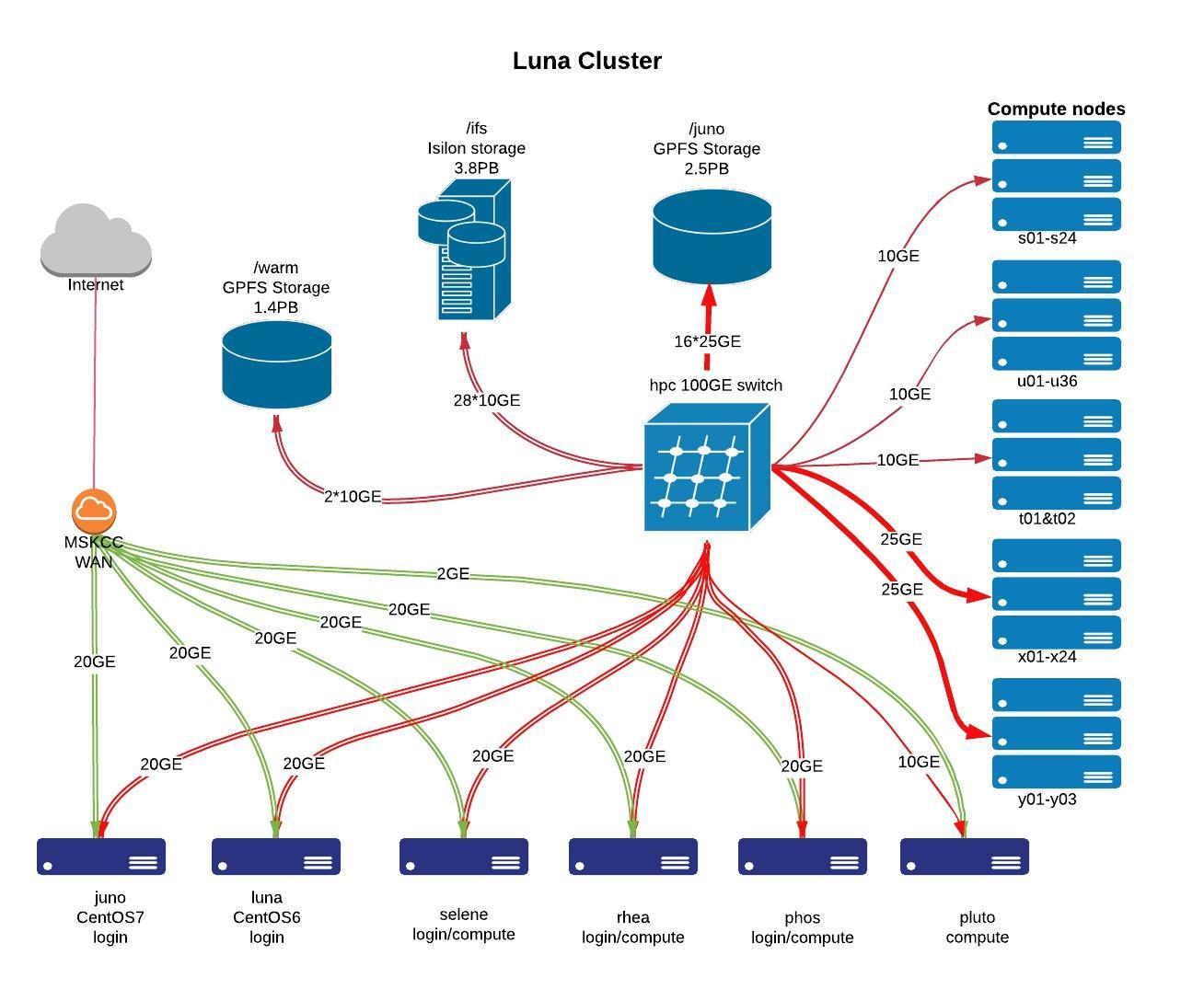
The Juno cluster is the successor to the Luna cluster. Juno has the same Isilon storage available as Luna. However, Juno’s CentOS 7 nodes have an additional 5.1PB GPFS filesystem. Most users have migrated to Juno. Luna is in the process of being retired.
Click here for a live view of system activity on Juno.
Compute
- CentOS 7 cluster login host: juno.mskcc.org
- 118 compute nodes:
| name | qty | model | CPU | #cores | RAM | #GPU | NVMe | GE | owner |
| ja01-ja10 | 10 | Supermicro SYS-6018R-TDW | 2 Xeon(R) CPU E5-2697 v4 @ 2.30GHz | 36 | 512 | – | 2x1T | 25 | CMO |
| jb01-jb24 | 24 | Supermicro SYS-6018R-TDW | 2 Xeon(R) CPU E5-2697 v4 @ 2.30GHz | 36 | 512 | – | 2T | 25 | Shah |
| jc01-jc02 | 2 | Supermicro SYS-7048GR-TR | 2 Xeon(R) CPU E5-2697 v4 @ 2.30GHz | 36 | 512 | 4 GeForce RTX 2080 Blower | 2T | 25 | Shah |
| jd01-jd04 | 4 | Supermicro SYS-6018R-TDW | 2 Xeon(R) CPU E5-2697 v4 @ 2.30GHz | 36 | 512 | – | 2T | 25 | Papaemm |
| ju01-u36 | 36 | HPE DL160G9 | 2 Xeon(R) CPU E5-2640 v3 @ 2.60GHz | 16 | 256 | – | – | 10 | CMO |
| jv01-jv04 | 4 | HPE DL160G9 | 2 Xeon(R) CPU E5-2640 v3 @ 2.60GHz | 16 | 256 | – | – | 10 | Offit |
| jw02 | 1 | HPE DL160G9 | 2 Xeon(R) CPU E5-2640 v4 @ 2.40GHz | 16 | 256 | – | – | 10 | CMO |
| jx01-jx34 | 34 | Supermicro SYS-6018R-TDW | 2 Xeon(R) CPU E5-2640 v4 @ 2.40GHz | 20 | 256 | – | 2x2T | 25 | CMO |
| jy01-y03 | 3 | Supermicro SYS-6018R-TDW | 2 Xeon(R) CPU E5-2640 v4 @ 2.40GHz | 20 | 512 | – | 2x2T | 25 | CMO |
Compute
- CentOS 6 cluster login host: luna.mskcc.org
- Additional compute servers: selene.mskcc.org, rhea.mskcc.org, phos.mskcc.org, & pluto.mskcc.org
- 25 compute nodes:
| name | qty | model | CPU | #cores | RAM | #GPU | NVMe | GE | owner |
| s01-s21 | 21 | HPE DL160G8 | 2 Xeon(R) CPU E5-2660 0 @ 2.20GHz | 16 | 384 | – | – | 10 | CMO |
| t01-t02 | 2 | HPE DL580G8 | 4 Xeon(R) CPU E7-4820 v2 @ 2.00GHz | 32 | 1536 | – | – | 10 | CMO |
| w01 | 1 | HPE DL160G9 | 2 Xeon(R) CPU E5-2640 v4 @ 2.40GHz | 16 | 256 | – | – | 10 | CMO |
Storage
/juno (CentOS 7 only): 2.6PB, GPFS v5.1, 4*NSD servers, 4*NetApp E5760, connected at 16*25GE
- /warm (non-compute): 1.5PB, GPFS v5.1, 4 * NSD Servers, 4 * Dell MD3460, connected at 4*10gbps
3.5PB, 28-node Isilon cluster. connected at 28*10GE
- /home — 100GB limit – for scripts only, no huge files, frequent mirrored backup
- /ifs/work — fast disk, less space, for active projects
- /ifs/res — slower disk, more space, for long-term storage of sequence data
- /ifs/archive — read-only – GCL FASTQ
- /opt/common — popular third-party programs
- /common/data — data, genome assemblies, GTFs, etc.
Network
Arista 7328x 100gbps switch
- approximately 600 TB of Isilon storage (mirrored in two data centers)