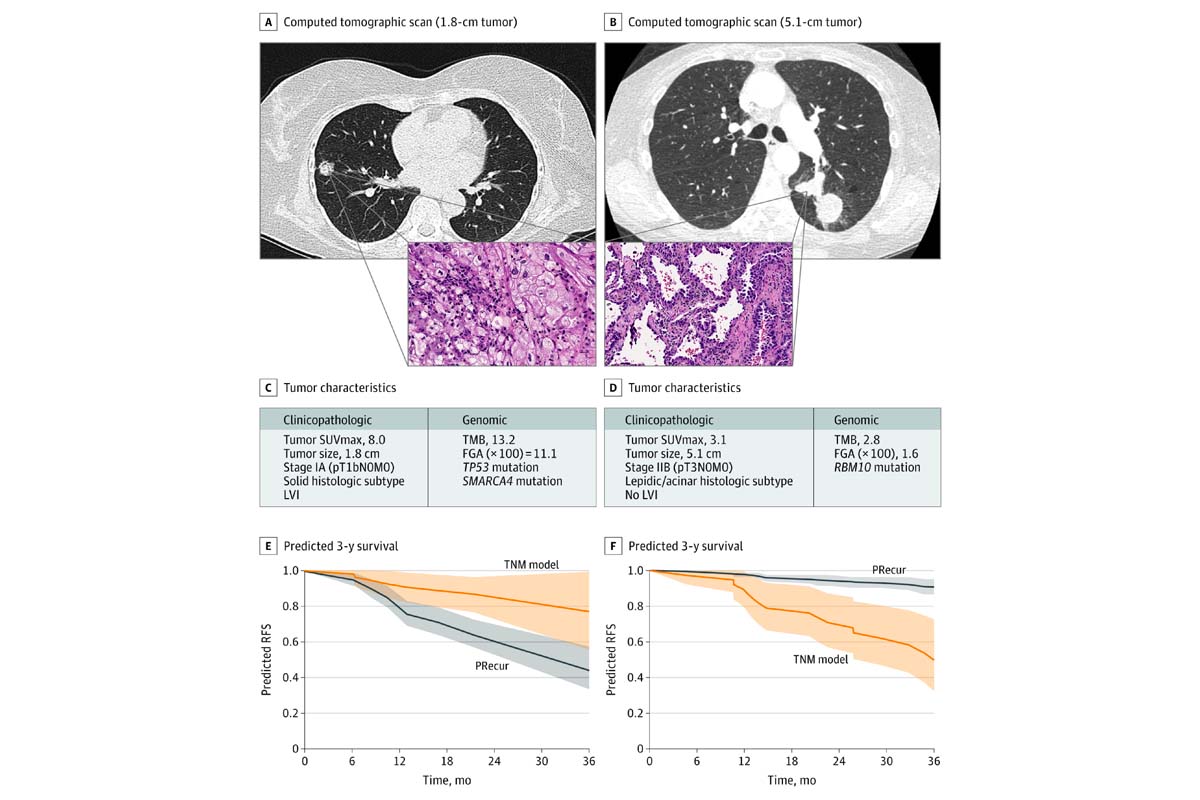
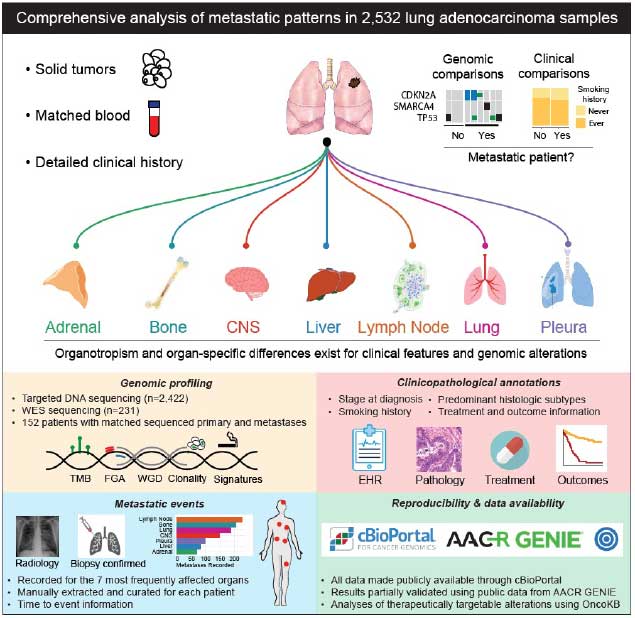
Despite complete surgical resection of stage I-III lung cancer, tumor recurrence develops in up to 50% of patients. Following recurrence, survival is poor, with five-year survival rates as low as 30% even with adjuvant cytotoxic chemotherapy for high-risk populations. The current rationale for recommending adjuvant therapy relies solely on TNM (tumor, node, and metastasis) classification, and more recently PD-L1 expression, and is largely agnostic to clinicopathologic and tumor genomic factors. As a result, survival benefits are modest at best when using the TNM model to select patients for adjuvant therapy. With the recent explosion of next-generation sequencing, our lab is rigorously investigating associations between tumor genomics, recurrence, and disease-free survival (DFS) in patients with early-stage lung adenocarcinoma (LUAD). In addition to a gene-centric approach, we have performed oncogenic pathway analyses to ascertain if these analyses can supplement existing TNM indicators for prognosis and prediction of recurrence. Using our large genomically annotated LUAD database, we were able to demonstrate unique differences in tumor genomics and mutational signatures across LUAD histologic subtypes. We have also identified the KRAS G12C mutation as a significant negative prognostic variable following complete resection of early stage LUAD4. We are interested in the prognostic abilities of tumor mutation burden (TMB), as well as the fraction of genome (FGA) - a measure of the percentage of the genome affected by gains or losses in copy number - and their association with LUAD recurrence. We developed PRecur, a novel computational machine-learning model that integrates genomic and clinicopathologic factors, to predict DFS in our MSK LUAD cohort (see figure below) and found that incorporation of both genomic and selected high-risk clinicopathologic variables is superior to TNM classification in predicting recurrence following surgery. We believe that continued investigation of tumor genomics in early-stage lung cancer has the possibility of moving the field in the direction of more personalized therapeutic decision making. Most recently, we analyzed thousands of LUAD samples to identify clinicopathologic and genomic features associated with metastasis, metastatic burden, organotropism, and metastasis-free survival. We found that inactivating mutations of TP53, SMARCA4, and CDKN2A correlate with a site-specific shorter time to metastasis. We actively collaborate with the Marie-Josée and Henry R. Kravis Center for Molecular Oncology at MSK, the Nikoulaus Schultz Laboratory, the Marcin Imielinski Laboratory, as well use MSK OncoKB®, MSK-IMPACT™, and related bioinformatics platforms.